Not many pre-retirement age professors abandon their running laboratories for a sabbatical to start anew as a young investigator in a different country. Yet this is what the 60 year old cancer researcher Carlos López-Otín did. This ERC-funded professor of Biochemistry and Molecular Biology left behind his 36-member-strong “Degradome” lab at the University of Oviedo in Spain and moved in with his collaborator in Paris, France, another degradome (autophagy and apoptosis) researcher Guido Kroemer.
That happened after my reporting uncovered numerous instances of inappropriate research data handling and publication from the Lopez-Otin lab, following his 2017 Mentoring Award issued by Nature. Who is going to mentor his over two dozens of postdocs, PhD and undergraduate students in Oviedo now, is anyone’s guess. But Lopez-Otin is now Professeur des universités-praticien hospitalier (PU–PH) at the Centre de Recherche des Cordeliers (CRC), which Kroemer is deputy director of.
The issues with Lopez-Otin’s research publications were mostly about re-used loading controls, retouched images and original data which did not match the published figures. It pales however when compared to what Kroemer himself has published, and this post will present some of that PubPeer evidence of duplicated gel bands. Maybe this is why Lopez-Otin moved in with Kroemer: research integrity is a relative issue, especially in France. Kroemer even signed that notorious CNRS Stalinist letter in defence of data manipulations and against whistleblowing and critical journalism.

Lopez-Otin departure from Oviedo was reported by the Spanish newspaper La Nueva Espana already in July 2018:
“I do not know when Carlos Lopez Otín will return to the University of Oviedo”, said yesterday the rector, Santiago García Granda, who considers the stay of Professor of Molecular Biology at the University Pierre and Marie Curie in Paris as a natural matter. “Moving is a normal thing”. […]
The temporary departure of Otín, García Granda said,is a part of the normal trajectory of the researchers: “He asked and was granted,” said Rector, who said that the professor “went alone” to work with one of his Gallic colleagues”.
The Gallic host of this refugee from Hispania was not named, but it is most obviously Kroemer. Lopez-Otin’s is associated with Kroemer’s department Apoptose, cancer, immunité at the CRC. I tried to find out what happened with Lopez-Otin’s fresh ERC grant of €2.5 million, awarded for 2017-2022, and whether the money stayed back in Oviedo or travelled with its owner to France to meet Kroemer. But unfortunately ERC does not communicate with me, and these EU funders of elite science do not seem to believe in research integrity anyway. In any case, ERC million-heavy grants are known to open hearts and minds, only the eyes stay closed, strangely.
My first article about Lopez-Otin appeared in December 2017, on the occasion of his Nature Mentoring Award, it described the PubPeer evidence of data manipulations in his papers. Then, in May 2018, my regular contributor Smut Clyde, discovered a “perennial Northern Blot“, which appeared in no less than 23 different papers from Lopez-Otin lab, over a period of 12 years. When my last article from August 2018 addressed the mismatch between the original research data and the presented results in a Nature Cell Biology paper, Lopez-Otin was already hiding in Paris with Kroemer, appreciating the French culture, cheeses and the country’s special variety of institutionalised research fraud.
That elite journal publication earned on October 4th an editorial note:
“Editor’s Note: We would like to alert readers that the reliability of data presented in this manuscript has been the subject of criticisms, which we are currently considering. We will publish an update once our investigation is complete.”
A key contributor to that study Soria-Valles et al Nature Cell Biology, 2015 was George Q Daley, Dean of Harvard Medical School and one of the most influential people worldwide in the field of stem cell research. After my May 2018 article and the comments it ensued, Daley set up a replication effort to reproduce the results he and Lopez-Otin published. At least two people were assigned to that project. The first author Clara Soria-Valles was not part of it though. She used to be a PhD student of Lopez-Otin’s, delegated to the Daley lab, where she remained as postdoc. In April 2018 however, Soria-Valles left Harvard, just when the post-publication peer review debate of her paper took off on PubPeer and the comment section of my site. Another paper of Soria-Valles’ was about to be submitted for publication to a stem cell society journal, but the study’s future and reliability are not sure, especially since all of its appointed authors left Daley lab by now.
For the Nature Cell Biology paper, Soria-Valles’ special contribution was the cell colony-counting technique, and those were the results which the commenters on my site and on PubPeer could make neither head nor tail of. Here are some examples, annotated by PubPeer users:
According to my source, Soria-Valles
“is not looking for her raw data to correct Nature Cell Biology, even though i) the journal published a comment last week; ii) Freije (another co corresponding of the paper) is contacting with all authors on the paper”
Jose Maria Perez Freije is professor at University of Oviedo, now doing the job of saving that paper without the help of Lopez-Otin or anyone else. Incidentally, Soria-Valles’ second co-author on her Nature Cell Biology paper is her fiancée Fernando Garcia Osorio, who also did PhD with Lopez-Otin and in this regard is responsible for a rather problematic paper of his own, Osorio et al Genes & Development 2012.
Did Soria-Valles and Osorio meet in a “creative Figure Preparation” course at Lopez-Otin lab? More on PubPeer.
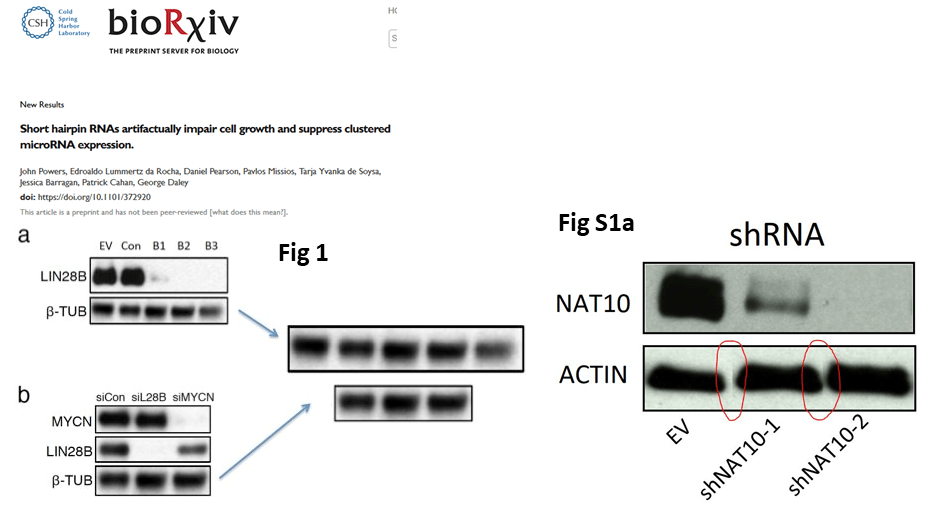
Daley’s lab now published a preprint, Powers et al 2018. Apparently, for Biorxiv one duplicated and one Photoshop-assembled loading control were good enough. Will a “better” version be produced for a proper journal, with impact factor?
It is a mess, and probably very stressful for everyone involved, and certainly the last thing Lopez-Otin, a hero of Spanish and international cancer research at the peak of his fame, needed. One fully understands why he left behind his huge lab and all the hassle in Oviedo and went to Paris: who wouldn’t? Kroemer is in this regard an excellent choice of a host. Lopez-Otin’s data integrity issues can seem as poppycock compared to what Kroemer and his life partner Laurence Zitvogel, based at the institute Gustave Roussy, dished out to the scientific community. Le Monde described this couple as an “explosive tandem”, it might just as well apply to their PubPeer record.
This is for example what Zitvogel and Kroemer produced in the highly impactful journal Nature Medicine, Apetoh et al 2007:

The relevant PubPeer thread elucidates that the blank gel fragments highlighted with colour circles look suspiciously similar up to the most minor details, not just in that Figure 3A. It would of course be interesting to know what the real western blots of those samples probed with those antibodies looked like. Or of their loading controls, because apparently also there things got duplicated out of every reasonable context.
Or how about this apparently duplicated set of Western blot bands in a Kroemer & Zitvogel co-production, published in EMBO Journal as Criollo et al 2010?
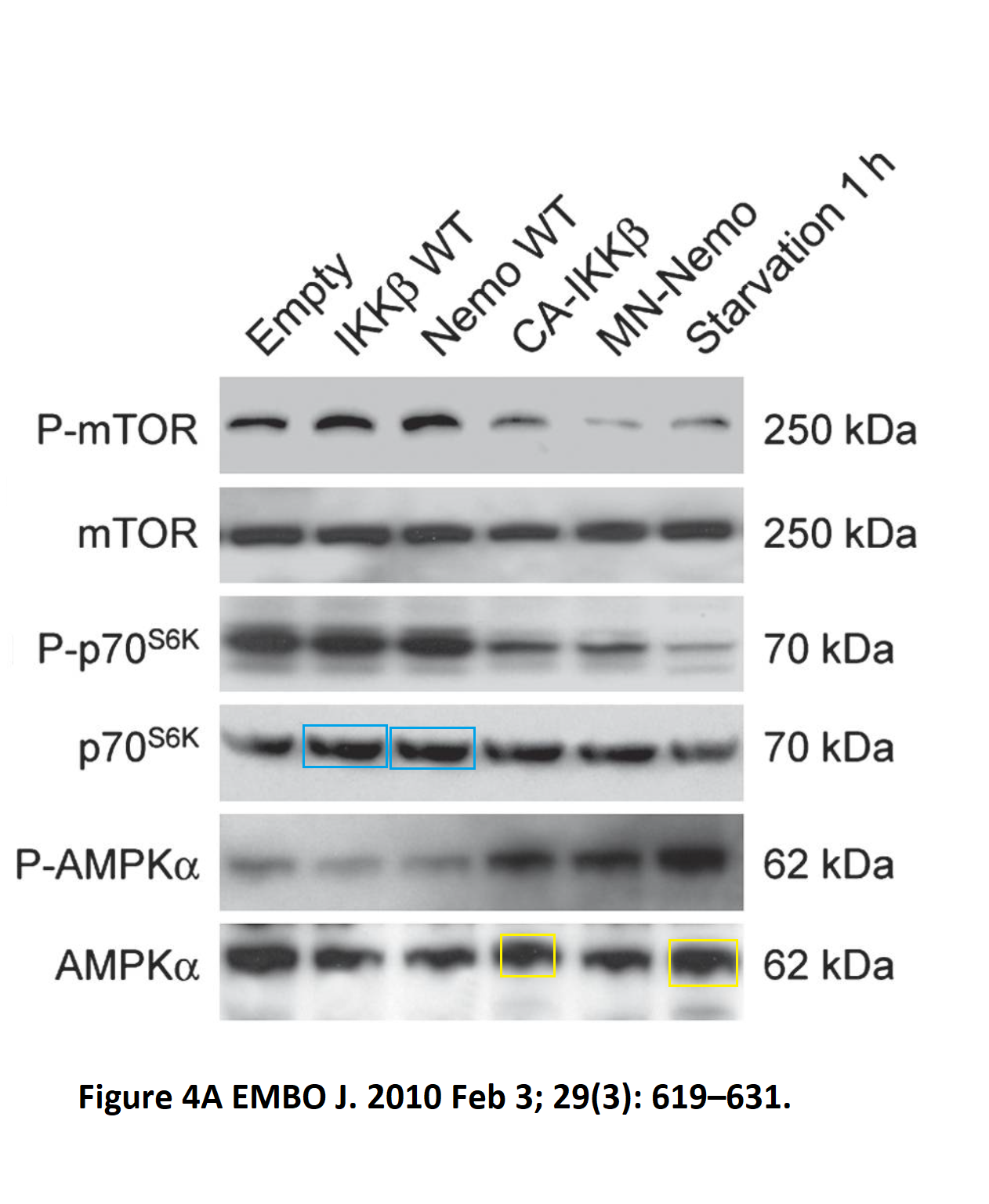
It just looks like laziness, or trolling. In any case, the lone PubPeer comment received no follow-up, from anyone. But there are many other papers on PubPeer, and some of those happened apparently before Guido met Laurence. Here is something from Kroemer’s bachelor times, again in EMBO Journal, Castedo et al 2002.
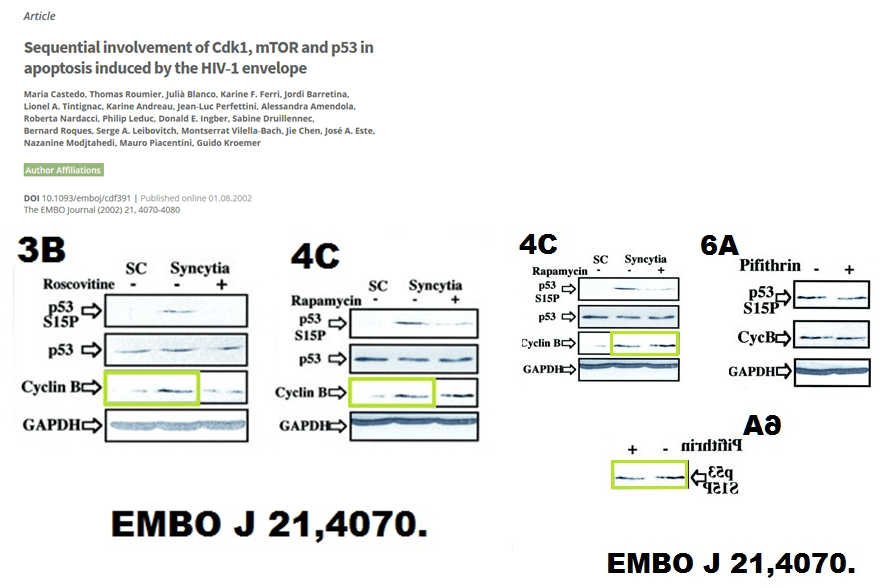
Kroemer is EMBO member, maybe this is why he is allowed to leave such beauties stand. EMBO Press seems to give a special leeway to EMBO members, like Olivier Voinnet or Maria Pia Cosma.
Here is another Kroemer-esque work of Photoshop art, same first author, Castedo et al J Exp Medicine 2001:
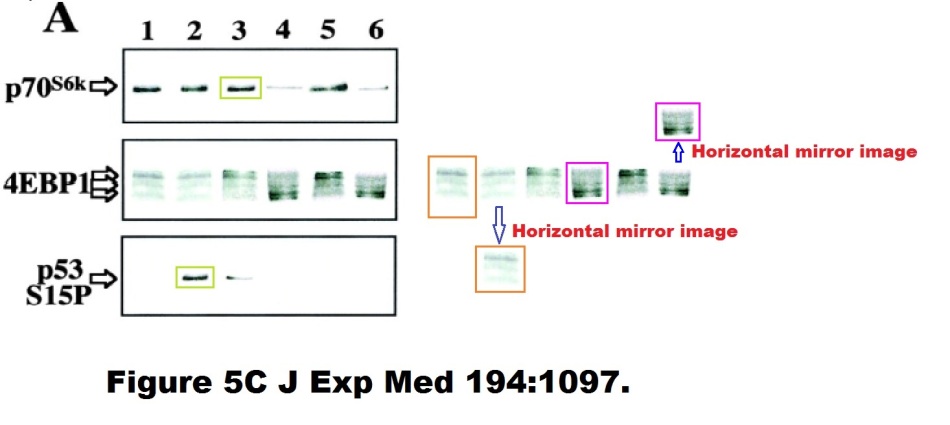
Enough for now, you got the message. If not, visit this Susin et al J Exp Medicine 1999 paper, showing some vintage western blot art by early Kroemer, or these beautifully corrected artworks Massard et al Oncogene 2006 or Zermati et al, Molecular Cell, 2007, where you can find some duplicated or even triplicated flow cytometry results for a change.
After all that, you probably will not be surprised that Kroemer was one of signatories of a recent Stalinist letter organised by CNRS. This letter defended data manipulations commited by CNRS chief biologist Catherine Jessus and the phony fraud-endorsing investigation by her friend and subordinate, the EMBO and Academie des Sciences member Francis-Andre Wollman, while calling for the heads of whistleblowers and critical journalists (more on that here).
What a company Lopez-Otin is now.

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism will be most appreciated!
€5.00


Why the hell do I continue to be a research scientist?
I struggle to get funding and to publish my findings, and
have realized that most of the papers I have been reading is
just bullshit.
When I see the “top scientist” in a flow of funding and
their manipulated and falsified data published in the top
journals I never have reached because my data in not
manipulated to be sexy enough for the sensational press
Nature, Science, Lancet etc, my thoughts go to all the
patients waiting for treatment based on their nonsense.
At the darkest hours when you realize that not only ETH, CNRS,
UCL, ICR, IDIBELL and ECR are rotten, but also WHO, then
you may seek comfort in some words from Churchill, a bit modifed though:
Even though most of Asia, and large tracts of Europe and USA have fallen or may fall into the grip of corrupt research institutions and funding agencies and all the odious apparatus of the publishers, we shall not flag or fail. We shall go on to the end. We shall fight in France, we shall fight in Spain and accross the Atlantic Ocean, we shall fight with growing confidence and growing strength in the air, we shall defend the clean and trusthworthy research, whatever the cost may be. We shall fight against the publishers, we shall fight at the research institutions, we shall fight in the meetings and conferences, we shall fight in the corridors and wherever we meet them; we shall never surrender, and if, which I do not for a moment believe, the island of honest were subjugated and starving, then our Empire of honest scientists armed and guarded by the evidence of fraud, would carry on the struggle, until, in God’s good time, the New clean World, with all its power and might, steps forth to the rescue and the liberation of the old corrupted World.
LikeLike
Dear Leonid, great article, thanks a lot. I think you are unveiling one of the key networks (in clear word, mafias) that is at the core of the corruption that affects European scientific research at all levels, including publishing and funding. In my opinion, it is not unreasonable Lopez-Otin went to Guido’s lab. To me, it looks like he went to the capo’s residence for protection. These kind of networks are really influential and vicious when they target someone. Many people know that one of the reasons why Lopez-Otin has an ERC grant is because he is part of a network of influence of which Kroemer is a highly influential member. These type of networks, which have proliferated in Europe, are composed of highly aggressive individuals who pretend to be “scientist leaders” and that gather huge groups of talented real scientists and postdocs to exploit them. They are not mentors. The only way these “scientists leaders” have to be successful is to destroy all the honest research scientists they have around and to build networks with their alikes to lobby EU politicians or young female journal editors. Many know that ERC is one such network. Lopez-Otin and Kroemer are alikes, although the former is at a lower level. By the way, Kroemer is not “Gallic”, is Austrian-Spanish. He got his PhD in 1992 at the Complutense University of Madrid. Amazingly, in his CV, he says he was group leader at the well known Center of Molecular Biology of Madrid (Spain), two years before his PhD!. This is what this kind of people consider “Excellent creative science cvs”.
https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=2&cad=rja&uact=8&ved=2ahUKEwjTt4euipDeAhXHK8AKHaxnDK4QFjABegQICBAC&url=http%3A%2F%2Fbrupbacher-foundation.org%2Ffileadmin%2Fdownloads%2FCancer_cell_stress_and_death_-_cell-autonomousand_immunological_considerations.pdf&usg=AOvVaw0tBQ19OsWE7u1DBPN1Qd-8
These type of “scientist leaders” have now learned and they are laughing out loud. Now Kroemer recommends matured cheese and expensive wine to cure cancer. Lopez-Otin stopped showing westerns and switched to next generation sequencing (NGS) and bioinformatics. He now keeps publishing in high impact journals (Nature, Blood). The advantage of big data NGS results is that you get nice figures that it is almost impossible to check whether they are fabricated or not or whether they have any relationship with reality. NGS is a mine gold for ex-Photoshop experts. Perhaps they got together to learn and improve these new sofisticates NGS tricks.
LikeLike
“individuals who pretend to be “scientist leaders””
2018 Pillar of “Gene Therapy” prize : Cancer Gene Ther. 2001 Apr;8(4):308-19.
Cancer Gene Ther. 2001 Apr;8(4):308-19.
Herpes simplex virus thymidine kinase/ganciclovir-induced cell death is enhanced by co-expression of caspase-3 in ovarian carcinoma cells.
McNeish IA1, Tenev T, Bell S, Marani M, Vassaux G, Lemoine N.
Author information
1
ICRF Molecular Oncology Unit, Imperial College School of Medicine, Hammersmith Hospital, London, UK.
Figure 5.
Figure 1.
Figure 6D.
Senior author presently: https://www.bci.qmul.ac.uk/en/staff/item/nick-lemoine
Director Barts Cancer Institute, Queen Mary Uiniversity London.
First author presently: http://www.imperial.ac.uk/people/i.mcneish
Chair of oncology, Imperial College, London.
LikeLike
All of these explains my lack of luck in research…because of this Hector Peinado will be keep invited as keynote speaker and others like Bruno Costa da Silva will get PI positions at Champalimaud Foundation after publishing two wonderfully pubpeer commented Nature papers
LikeLike
Ana, can you tell more about what happened to you and what your experience in science has been?
LikeLike
Meet up at conferences where these guys frequently are and ask if they can explain all the questionable data posted on Pubpeer and why they are not retracting their invalid conclusions. Of course in plenary sessions. Let’s make the most embarrassing to those who are ruining the science. Let them feel ashamed, although these people most probably are not able to feel shame.
LikeLike
Although I have very short time I will try to prepare a few questions for Hector Peinado at the UKEV2018 on December 11 in Sheffield
I can add the question: if their data was so good why I had to receive an email from Champalimaud Foundation cc David Lyden/Fatima Cardoso frightening me and asking me to delete my work data which compromised them and also why Dr Inglis received an email from them also requesting deleting my pre-print from BioRXiv
LikeLike
Did you publish these emails? Those threats are definitely actus reus and proofs that these individuals are not scientists.
LikeLike
Curiously please do correct me if I am wrong but Hector Peinado is not anymore among keynote speakers for the UKEV2018…..
LikeLike
Love the play on Vogel in the title!
LikeLiked by 1 person
To aiaas,
Here it goes the first email:
From: Joaquim Teixeira joaquim.teixeira@fundacaochampalimaud.pt
Sent: Friday, September 30, 2016 11:50 AM
To: Ana Pedro
Cc: Tiago Rosa Mendes; Sofia Bragança; David Lyden; Fatima Cardoso
Subject: RE: Thank you RE: Statement
Dear Ana,
It came to my attention that you intend to submit an abstract to a meeting that will take place in December and there present a poster that will include at least part of the data belonging to the Champalimaud Foundation and Cornell University.
We strongly advise you not to do so. We also strongly advise you to immediately cease making use of any data or information belonging to the Champalimaud Foundation. Such action violates formal agreements between you and the Champalimaud Foundation. Furthermore, it will limit, harm and/or jeopardise the legitimate interests of the Champalimaud Foundation. We will thus be forced to take legal action against you, including to obtain injunctive orders against your actions.
Please allow me to reiterate the following, as communicated to you and acknowledged by you on the 27th of April of 2016:
First: “All information, including all scientific outputs directly or indirectly generated by you, using those samples, under this project, belong to the Champalimaud Foundation. They do not belong to you and you are not to withhold any information that belongs to the Champalimaud Foundation without our express consent. You currently do not have our consent.”
Second: “You are not authorised to keep, hold, use or modify any information, including any scientific data generated directly or indirectly from the patients’ samples. If you are still in possession of any such information or data and you have not done so already, you are to return all copies immediately to the Champalimaud Foundation.”
Furthermore, on the 27th of April you also informed us that “As soon as possible [you] will deleted the files from the dropboxes (you get check [sic] later on) and I don’t have more any files belonging to CF.”
It came to our attention that not only you are perhaps still in possession of data belonging to the Champalimaud Foundation, including raw mass spec data, but also that you may have sent said information to others, as well as shared links to cloud storage where you maintain and control copies of our data.
That would mean that you never deleted the files. That would also mean that you have made available to third parties data that belongs to the Champalimaud Foundation without our authorisation. Do you confirm these allegations?
I must warn you that, if this is true, your actions have and/or may result in grave harm to the Champalimaud Foundation. We will thus have no choice but to move swiftly and take further legal action against you, as well as to seek remedy and compensation.
The Head of our Legal Department, Dr. Tiago Rosa Mendes, in copy, has been notified. You should expect to hear from our Legal Department very soon.
Best regards,
Joaquim
LikeLike
And the second email:
rom:
Inglis, John inglis@cshl.edu
to:
“anapedrolaboratories@gmail.com” anapedrolaboratories@gmail.com
cc:
“fatimacardoso@fundacaochampalimaud.pt” fatimacardoso@fundacaochampalimaud.pt,
“joaquim.teixeira@fundacaochampalimaud.pt” joaquim.teixeira@fundacaochampalimaud.pt,
“irm2224@med.cornell.edu” irm2224@med.cornell.edu,
“dcl2001@med.cornell.edu” dcl2001@med.cornell.edu,
biorxiv biorxiv@cshl.edu
date:
Mar 10, 2018, 3:11 AM
subject:
Your bioRxiv submission
Dear Dr Pedro:
We have received the message below from colleagues at Weill Cornell Medicine NY and the Champalimaud Foundation, Lisbon. They make allegations concerning the manuscript you posted to bioRxiv in October 2017.
I would appreciate receiving your response to these allegations. At bioRxiv, we will take the actions we feel are appropriate when we receive your reply.
Sincerely,
John Inglis
Dr John R Inglis
Co-Founder, bioRxiv (biorxiv.org)
Executive Director and Publisher
Cold Spring Harbor Laboratory Press (cshlpress.org)
1 Bungtown Road
Cold Spring Harbor
NY 11724
Office:516-422-4005
Email:inglis@cshl.edu
Twitter: @JohnRInglis
From: Irina Matei [irm2224@med.cornell.eduirm2224@med.cornell.edu]
Sent: Wednesday, March 07, 2018 6:25 PM
To: biorxiv feedback
Cc: David C Lyden; Mark T. Adams; fatimacardoso@fundacaochampalimaud.ptfatimacardoso@fundacaochampalimaud.pt; ‘joaquim.teixeira@fundacaochampalimaud.pt’joaquim.teixeira@fundacaochampalimaud.pt‘
Subject: Request for manuscript removal from your repository
Dear BioRxiv Editors,
I am writing on behalf of Dr. David Lyden of Weill Cornell Medicine, NY, NY, USA and Dr. Fatima Cardoso of the Champalimaud Foundation, Lisbon, Portugal.
It has come to our attention that a manuscript by Dr. Ana Pedro entitled “Identification of potential blood-derived extracellular vesicles biomarkers to diagnose and predict distant metastases in ER+ breast cancer patientshttps://urldefense.proofpoint.com/v2/url?u=https-3A__www.biorxiv.org_content_early_2017_10_12_202291&d=DwMFAw&c=mkpgQs82XaCKIwNV8b32dmVOmERqJe4bBOtF0CetP9Y&r=0jX5MDh43jzUfm5EA1F-OV8VuRZelKG5yLnmapw3osk&m=-A0OuUja0HCDI-iBkmmHweIC67WH5u8w9STaJHYygSk&s=vZQPOaLFzY2A2v0_Q_LwSykfH21fGZHUqWmzmXyqU-s&e=” had been deposited to your repository and made available to the scientific community. Dr. Pedro does not own these data and does not have permission from the rightful owners, Drs. Lyden and Cardoso, to publish it. Please find attached a letter from Drs. Lyden and Cardoso, detailing the situation.
Given the reputation of the Cold Spring Harbor Publishing and that bioRxiv upholds high publication standards by reserving the right to identify and remove any articles that contain plagiarized material or describe experimental work that is not performed in accordance with the relevant ethical standards for research using animals or human subjects, we respectfully request that the article by Ana Pedro be removed from your repository as it violates these policies.
Thanking you in advance,
Best regards,
Irina Matei, PhD
Laboratory of Dr. David Lyden
Weill Cornell Medicine
Children’s Cancer and Blood Foundation
Departments of Pediatrics, Cell & Developmental Biology
Drukier Institute for Children’s Health and Meyer Cancer Center.
413 East 69th Street, Belfer Research Building
12th Floor, Room BB1227/BB1250
New York, NY 10021
Phone: 646-962-4878
Fax: 646-962-0574
irm2224@med.cornell.eduirm2224@med.cornell.edu
Kind regards,
Ana Pedro
LikeLike
What does Prof. David Lyden is really fearing?
This: https://elifesciences.org/articles/39944 or this: https://pubpeer.com/publications/B1C028796E9137389615799320FB1C or even this: https://pubpeer.com/search?q=david+lyden?
Or even concerns about the PMN…?
LikeLike
Surprise!: ERC Advanced Grant Panel 2017: LS2 panel on Genetics, Genomics….panel member: Manel Esteller (non-confessed scientific counterfeiter). ERC Advanced Grant Panel 2016: LS4 panel on Physiology. Panel member Mariano Barbacid. Who reviewed ERC Lopez-Otin application on aging? Perhaps a coincidence:
https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=2ahUKEwi60-Th46HeAhXIyqQKHStpDmYQFjAAegQICRAC&url=https%3A%2F%2Fwww.fundacionlilly.com%2Fes%2Factividades%2Fsimposio-cientifico%2Feasset_upload_file774_68001_e.pdf&usg=AOvVaw2fwUWANQGa4tBtiQTiBYIc
LikeLike
Pingback: Olivier Voinnet, the new Dreyfus? – For Better Science
“Another paper of Soria-Valles’ was about to be submitted for publication to a stem cell society journal, but the study’s future and reliability are not sure, especially since all of its appointed authors left Daley lab by now.”
What is this paper? Is this published already?
LikeLike
I know which journal and I informed the Editor-in-Chief now.
LikeLike
PhD mentor of George Q. Daley is Nobel laureate David Baltimore who found NF-kB. Quite interestingly, Soria-Valles and Lopez-
Otin brought brilliant idea for Daley to put stem cells, aging and NF-kB in turmoil. Their main claim of aged fibroblasts resist to reprogramming has been challenged by a new pre-print from Anne Brunet lab. And causative role of NF-kB in reprogramming was not verified. And waht’s more, unlike minimal tiny error bars in NCB paper, inter-group variability of reprogramming efficiency was way bigger in this pre-print, that follows the nature of this experiment. “We did not observe a significant change in the mean reprogramming efficiency of cells from older individuals (Fig. 1d-e). In contrast, there was a significant increase in the individual-to-individual variability of reprogramming efficiency in cells from old donors, with cells from some old individuals reprogramming either better or worse than those from young individuals” https://www.biorxiv.org/content/early/2018/10/19/448431
LikeLike
“In April 2018 however, Soria-Valles left Harvard” Is she still payed salary? I still see her name on George Daley’s lab website. It’s been more than 6 months since she left. https://daley.med.harvard.edu/people
Is it common academia practice to pay NIH (or fellowship) money to keep someone’s absent seat for long? I guess the Daley is the most competitive lab to join in stem cell field. Many enthusiastic, bright students and postdocs should be applying. Totally non-sensical that the empty seat has been blocking potential new comer for long…
LikeLike
This seems more a forum of Asturian marujas than a scientific forum
LikeLike
Soria-Valles is an EMBO (ALTF 1240-2015) fellow.
Daley’s recent fundings:
US National Institute of Diabetes and Digestive and Kidney Diseases (R24DK092760),
the National Institute of Allergy and Infectious Diseases (R37AI039394),
the National Heart, Lung, Blood Institute Progenitor Cell Biology Consortium (UO1-HL100001) ,
the Progenitor Cell Translation Consortium (UO1-HL134812),
Alex’s Lemonade Stand Foundation,
the Doris Duke Medical Foundation,
NIH NIDDK R24-DK49216,
NHLBI R01HL04880,
NIH R24OD01787001, and
the Manton Center for Orphan Disease Research
Yep, they have enough money which is totally fine to dispense to warm up empty bench.
LikeLike
Apparently, PubPeer closed comments to Soria-Valles Nature Cell Biology article. But readers are still raising new concerns in another thread. https://www.pubpeer.com/publications/0DFD5A1F3303958FBBE3D562F455AE
New claim; Their fibroblast reprogramming experiments actually seem merely replating existing iPSCs.
“In addition, this NCB paper has several points I cannot make head and tail with. Reprograming efficiency using 4 independent retroviral vectors are so high and consistent. I endorce above section critisisms of too tiny error bars. One needs lentiviral polycistronic vector to achieve the results, but if I see Methods section, “retroviral vectors expressing human OCT4, SOX2, KLF4 and c-MYC” . Indeed, inter-group variability of reprogramming efficiency was way bigger in Brunet’s pre-print, that follows the nature of this experiment. Time course of reprogramming was too short. When I saw figure 3e, I was surprised that TRA-1-60+ colonies were formed within 1 week, and colony numbers reached plateau by 2 weeks. If one reprogrammed human fibroblasts, it actually takes longer to be able to see TRA-1-60+ colonies. The timecourse shown in figure 3e is more likely that of replating existing iPSCs, not reprogramming fibroblasts.”
LikeLike
Concerns over Soria-Valles’ Nature Cell Biology paper continue… New PubPeer comments.
https://pubpeer.com/publications/0DFD5A1F3303958FBBE3D562F455AE#5
Questionable statistics.
Raw data actually showed opposite conclusion.
LikeLike
Capture of The Daley Lab members as of Nov 11 2018. Clara Soria-Valles, Ph.D. Postdoctoral Fellow.
LikeLike
Here is an abstract of Stem Cell Community Meeting (ISSCR 2017@Boston) Soria-Valles et al. presented and are going to submit to its associated journal Stem Cell Reports for publication.
INTEGRATION-FREE SYSTEM FOR GENERATION OF HEMATOPOIETIC STEM AND PROGENITOR CELLS FROM HUMAN PLURIPOTENT STEM CELLS
Soria-Valles, Clara1, Sugimura, Ryohichi1, Kumar Jha, Deepak1, Lummertz da Rocha, Edroaldo1 and Daley, George2
1Boston Children’s Hospital, Boston, MA, USA, 2Stem Cell Program, Boston Children´s Hospital, Boston, MA, USA
The generation of hematopoietic stem cells (HSCs) from human pluripotent stem cells (PSCs) constitutes a valuable tool with promising applications for research and therapy. However, derivation of HSCs with in vivo long-term engraftment and multi-lineage potential remains elusive. We have described a combinatorial approach, based on the directed differentiation of hemogenic endothelium (HE) and transduction with five transcription factors (TF) (RUNX1, ERG, LCOR, HOXA5 and HOXA9) expressed in lentiviral vectors that allowed the conversion of human PSCs into hematopoietic stem and progenitor cells (HSPCs). The resulted cells exhibited long-term and multi-lineage hematopoietic capabilities when injected into irradiated immune-deficient mice. Despite this proof of principle, the engineered cells have a limited self-renewal capacity due to the integration of the transgenes and are still molecularly distinct from bona fide HSCs. Thus, in an attempt to achieve bona fide HSCs and make them safer for future therapeutic interventions, we have established integration-free systems that have shown comparable efficiency to the previously developed lentiviral strategy through in vitro and in vivo experiments. Therefore, this new method may overcome some limitations of the lentiviral approach and hold the key for future regenerative medicine advances in blood diseases.
http://www.isscr.org/docs/default-source/am17/64553-isscr-abstracts-proof-3-w-cover.pdf?sfvrsn=2
LikeLike
Look at this! PubPeer starts deleting comments on Soria-Valles Nature Cell Biology and Protocol Exchange.
Is this why Lopez-Otin moved to France, the physical location of PubPeer?
165 –> 154 comments. https://www.pubpeer.com/publications/836AB3A8AB4FD562A2D4CBFBF8ED18
6 –> 4 comments. https://www.pubpeer.com/publications/0DFD5A1F3303958FBBE3D562F455AE
LikeLike
Interesting discrepancy between Brunet’s pre-print and Lopez-Otin’s NCB.
Gathering attention on Pubpeer.
https://pubpeer.com/publications/ED47ED7138B4141C62F0AFAA6EA15A#4
“Reprogramming efficiency in old fibroblasts; – consistently impaired n=2 (Soria-Valles, 2015), – variable n=53 (Mahmoudi, 2018).”
“Although NF-kB signature was detected in expression profile, actual protein blot did not indicate it. In contrast, Soria-Valles et al. showed very thick band of blots. How it comes?”
LikeLike
Another discrepancy between Brunet’s pre-print and Lopez-Otin NCB.
I do curious if progeria- and elderly-iPSC lines are available from Lopez-Otin lab to validate their results.
“Aged fibroblast-derived iPSCs differentiate to three germ layers normaly in this report. In contrast, Soria-Valles’ NCB paper reported impaired differentiation capacity. Could be best to request their progeria-iPSC lines and carefully check it. ”
LikeLike
Why post-natal cells could not make iPSCs in Park et al., Nature paper, which was Dean Daley’s most important achievement?? Ironically, Soria-Valles clearly showed that the same lab can generate iPSCs from post-natal cells.
Reprogramming of human somatic cells to pluripotency with defined factors
Nature (2008) – 1 Comment
pubmed: 18157115 doi: 10.1038/nature06534 issn: 1476-4687 issn: 0028-0836
In-Hyun Park , Rui Zhao , Jason A. West , Akiko Yabuuchi , Hongguang Huo , Tan A. Ince , Paul H. Lerou , M. William Lensch , George Q. Daley
“It is striking to see no derivation of iPSCs from post-natal sources in three independent cell lines (Table 1). Conclusive that 4 factors cannot reprogram post-natal human cells regardless of tissues (foreskin, lung and dermis). In contrast, follow-up paper from the same senior author reported successful derivation of iPSCs from post-natal sources (Soria-Valles et al., 2015). How to explain this? Both studies used same reprogramming factors, OCT4, SOX2, KLF4 and MYC”
https://pubpeer.com/publications/A600E09BABFB5C7A28B86BBA6F8AD9
LikeLike
Issue in another career-building Nature paper from George Daley.
Duplicated image of teratoma…
Epigenetic memory in induced pluripotent stem cells
Nature (2010) – 1 Comment
pubmed: 20644535 doi: 10.1038/nature09342 issn: 1476-4687 issn: 0028-0836
K. Kim , A. Doi , B. Wen , K. Ng , R. Zhao , P. Cahan , J. Kim , M. J. Aryee , H. Ji , L. I. R. Ehrlich , A. Yabuuchi , A. Takeuchi , K. C. Cunniff , H. Hongguang , S. Mckinney-Freeman , O. Naveiras , T. J. Yoon , R. A. Irizarry , N. Jung , J. Seita , J. Hanna , P. Murakami , R. Jaenisch , R. Weissleder , S. H. Orkin , I. L. Weissman , A. P. Feinberg , G. Q. Daley
LikeLike
Park et al., Nature paper from Daley lab recieved more critiques from PubPeer readers.
“This paper proposed that Yamanaka factors are insufficient, and you need both hTert and large T antigen to fully reprogram cells. But no one uses hTert and large T antigen any longer in the field.”
“Their iPSCs were not fully reprogrammed. They still expressed and depended on transgenes, which is of concern. ”
https://pubpeer.com/publications/A600E09BABFB5C7A28B86BBA6F8AD9
LikeLike
Wow. It’s amazing. Every cell line has transgene turned on. Clear evidence of failed reprogramming.
LikeLike