Clare Francis recently investigated another set of French scientists, and posted it on PubPeer. One of them is the cancer researcher Anne Dejean- Assemat, Research Director Classe Exceptionnelle at INSERM and professor at the Institut Pasteur in Paris. The other is her German mentee Oliver Bischof, now group leader under Dejean’s leadership. It is very likely Bischof is being groomed to succeed Dejean when she retires. And to be fair, the way this Pasteur lab is run, he is definitely the right man for the job.
This is Dejean’s CV, as presented by her “Nuclear Organization and Oncogenesis” lab at Institut Pasteur:
“Anne Dejean is Research Director at INSERM, Professor at the Institut Pasteur and Head of the Laboratory of Nuclear Organization and Oncogenesis/INSERM U993. She graduated from Pierre et Marie Curie University in Paris and earned her PhD in Pierre Tiollais’ lab at the Institut Pasteur in 1983. Member of EMBO and of the French Academy of Sciences, she has received the Gagna and Van Heck Prize in 2003, the L’Oréal-UNESCO for Women in Science Awards in 2010, the Grand Prix INSERM in 2014 and the Sjöberg Prize in 2018. She was awarded two ERC Advanced Grants, in 2011 and 2018.”
I would like to show you how exactly Professor Dejean achieved so much. And yes, I know European Research Council (ERC) leadership doesn’t believe in research integrity, but still. The scientific community has a right to know who and how receives such exclusive public research funding, which honest scientists see routinely denied. Dejean’s new €2.5mn Advanced Grant “Deconstructing the role of SUMO on chromatin in cell identity and tissue repair” was awarded in 2018, and don’t worry, it will continue for the next 5 years excellently. On this, you can trust ERC. Carlos Lopez-Otin enjoys his ongoing grant despite retractions, mouse murder, admitted data fakery and cessation of research activities, and even proven the fraudster Maria Fousteri was free to use up all of €2mn to her pleasure, after the university notified the ERC.

Now, someone, or a group of someones in Dejean’s lab has been naughty. We cannot know who, but the evidence of their naughtiness is there. Let’s look at this almost 20 year old paper:
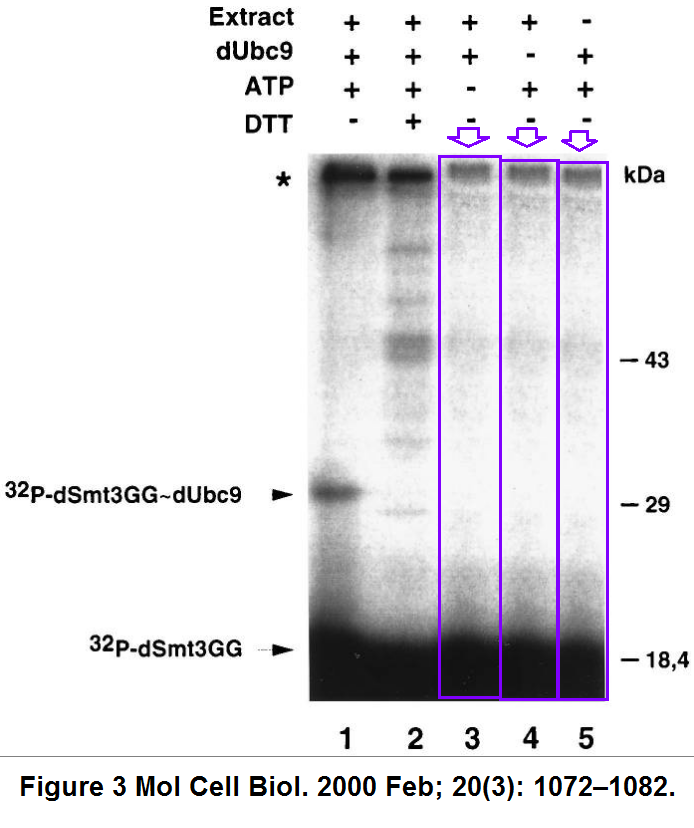
F. Lehembre, P. Badenhorst, S. Müller, A. Travers, F. Schweisguth, A. Dejean Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies Molecular and Cellular Biology (2000) doi: 10.1128/mcb.20.3.1072-1082.2000
Normally, gel lanes do not triplicate like this, unless the authors disagreed with their actual experimental results and decided prove them wrong, using The Photoshop, or in this case, probably the good old scissors and glue. Maybe the issue is serious enough for the journal to overrule the 6-year deadline strategy? After all, the journal MCB already had to correct a Dejean-coauthored paper from INSERM in Paris, for duplicated data (Erker et al MCB 2013).
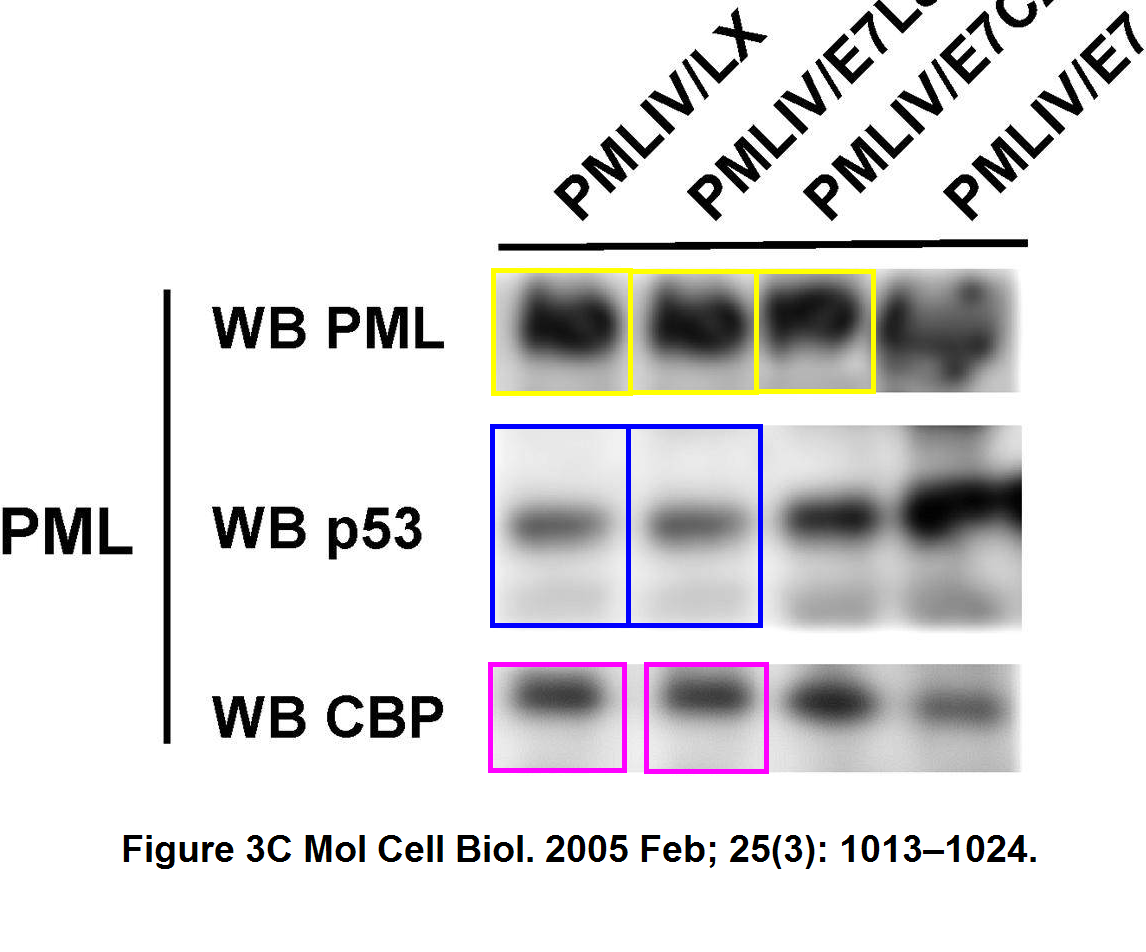
Or how does this happen, in the same journal, but 5 years later? Something horrible happened to Figure 3C in 2005, this time it was most likely the revolutionary biomedical research tool of Photoshop being applied:
O Bischof, K Nacerddine, A Dejean Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways Molecular and Cellular Biology (2005) doi: 10.1128/mcb.25.3.1013-1024.2005
How come 3 out of 4 bands of the top PML western blot look so similar, especially after a rotation? Also the two central E7 bands seem to be mirror images of each other, and it turned out, also the first two p53 bands are strangely similar, and the same applies for the CBP bands below.
How exactly did Dejean and Bischof plan to cure leukaemia with this technology? One year later, they published another paper with similar issues, where some of Bischof et al MCB 2005 data was apparently reused in excitingly new context. Luckily the new journal was Molecular Cell, this Elsevier outlet seems to have an unhealthy obsession of attracting and cultivating manipulated data.
O Bischof, K Schwamborn, N Martin, A Werner, C Sustmann, R Grosschedl, A Dejean The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis Molecular Cell (2006) doi: 10.1016/j.molcel.2006.05.016

Now, what do we have here? A set of 3 bands from Figure 2C of MCB 2005 which used to be p53, but then somehow became Rb, a very different protein. One of this trio of jumping bands grabbed another friend and cloned themselves again to become yet another protein, Cyclin D1 in Figure 4E of Molecular Cell. It is not helpful that the sample legend is different for every single instance. And since we are discussing the magical permutation of p53 into Rb protein, let’s look at Figure 4A:

The band even got hyper-phosphorylated while transforming itself from p53 to pppRb. There are more gems in that 2006 paper, have a look at Figure 6H:
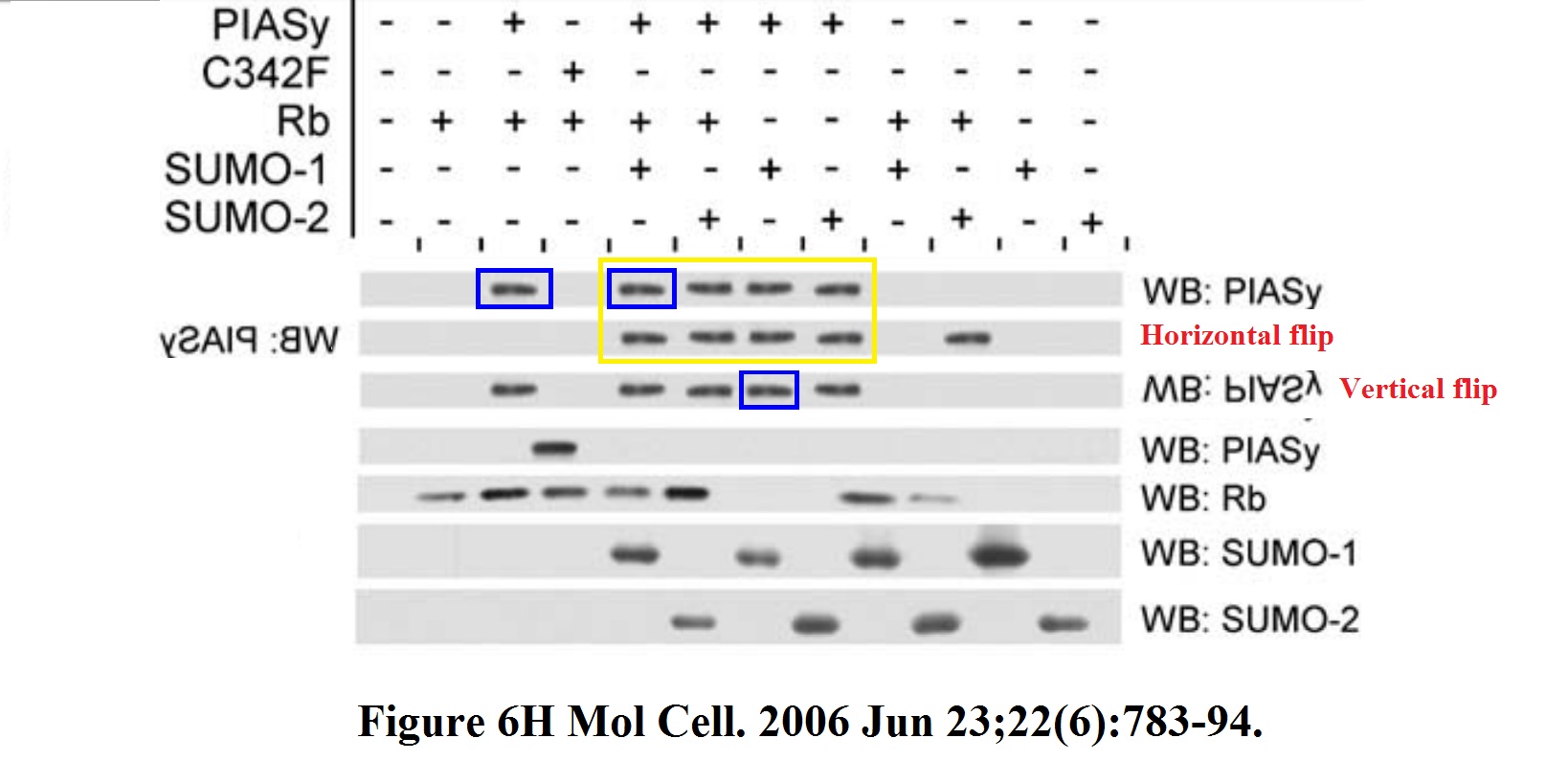
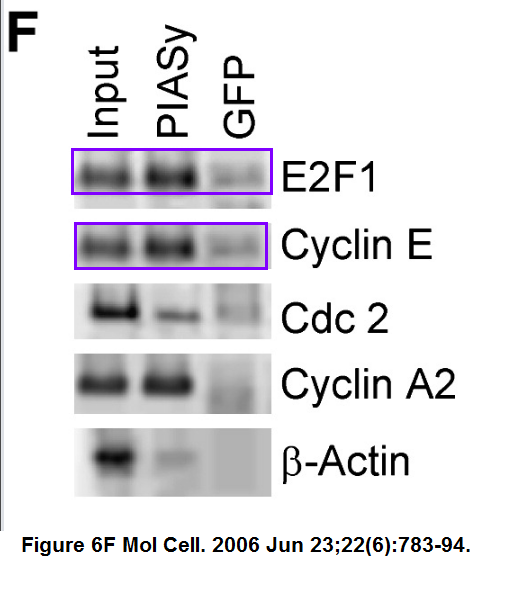
Can it be that top western blot panel shows the same band all over, copy-pasted with an occasional flip? Was someone taking the proverbial PIAS here? Also the Figure 6F has what Institut Pasteur experts will probably call “an inadvertent duplication”.
Now, Clare Francis did report all these and other issues to Molecular Cell editors, but it is rather like writing to Philip Morris to complain about someone smoking in a restaurant. This Cell Press journal chose to do nothing at all about fraudulent papers by former CNRS president Anne Peyroche, and rejected retraction requests in the case of Maria Fousteri.
Before the first author of this Photoshoppian masterpiece joined the Dejean lab in Paris, the German Oliver Bischof did a brief postdoc stint at the lab of Farzin Farzaneh at King’s College London. There was much to learn also: Farzaneh was recently investigated for data manipulation, but no research misconduct was found, only “poor research practices” which required Errata in 5 journals.
Between Farzaneh and Dejean postdoc, Bischof did a stint at the Berkeley lab of Judith Campisi, a star researcher in the field of cellular senescence. This collaboration moved the research area ahead with this important contribution:

O Bischof, S Galande, F Farzaneh, T Kohwi-Shigematsu, J Campisi Selective cleavage of BLM, the bloom syndrome protein, during apoptotic cell death The Journal of biological chemistry (2001) doi: 10.1074/jbc.m006462200
Now this cannot have happened by incident, no matter how drunk The Bischof might have been, because the upper and lower set of bands are supposed to have been on one solid gel. No way the upper 20/46 bands can accidentally become lower 20 set, flipped.
The shenanigans apparently continued in Paris. This last-author paper from Institut Pasteur Bischof proudly shows among his selected publications:
M Ogrunc, RI Martinez-Zamudio, P Ben Sadoun, G Dore, H Schwerer, P Pasero, JM Lemaitre, Anne Dejean, O Bischof USP1 Regulates Cellular Senescence by Controlling Genomic Integrity Cell Reports (2016) doi: 10.1016/j.celrep.2016.04.033
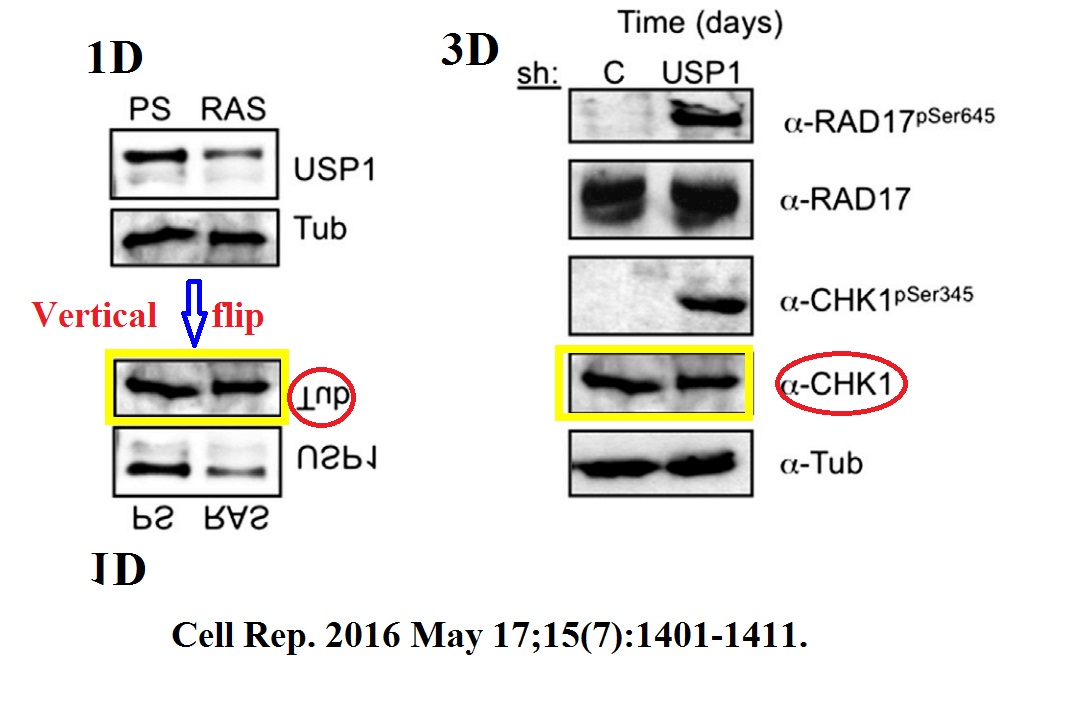
Now I should immediately declare that the first author Müge Ogrunc and I worked for several years together at the same DNA damage research small lab at IFOM in Milan, Italy. I tried to contact Müge about this copy-pasted and flipped CHK-1 western blot, but received so far no feedback. But then again, my former colleague Müge joined the Dejean-Bischof lab at Pasteur only for two years, 2015-2017, while the data integrity issues go way back there.
Dejean is now 62 and will probably retire in a couple of years, maybe when her new ERC grant ends. With Bischof, she prepared a worthy successor to take over her artisan craft business, which now started to conquer new sources of grant cash: ageing or senescence research. The general questionable research ethics in Dejean lab shine through in examples like this:
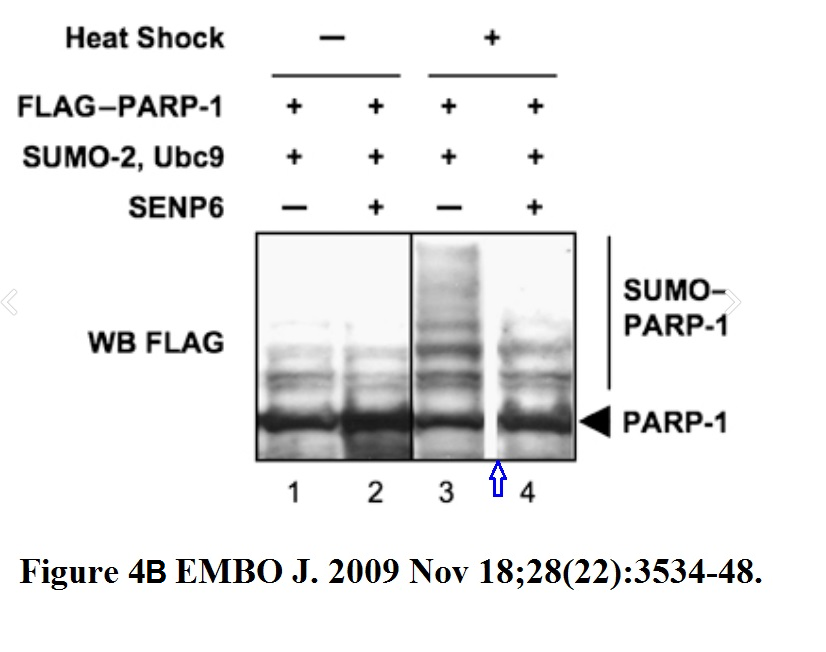
N Martin, K Schwamborn, V Schreiber, A Werner, C Guillier, XD Zhang, O Bischof, J Seeler, A Dejean PARP-1 transcriptional activity is regulated by sumoylation upon heat shock The EMBO Journal (2009) doi: 10.1038/emboj.2009.279
Now the paper is 10 years old, and the French and German authors might declare that they missed previous guidelines in biomedical publishing where gel splicing should be avoided, and where not possible, clearly indicated, because the guidelines were in English. But thing is, in Figure 4B they did provide a clear black vertical line to indicate splicing in one case, but they chose not to do this in other cases. Why? Because you cannot compare the signal of a + vs – treatment on a spliced gel. In brief, the authors bullshitted the EMBO editor, the peer reviewers and the scientific community while knowing perfectly well that what they did was not right.
Or how about another paper from same time, with same key authors:
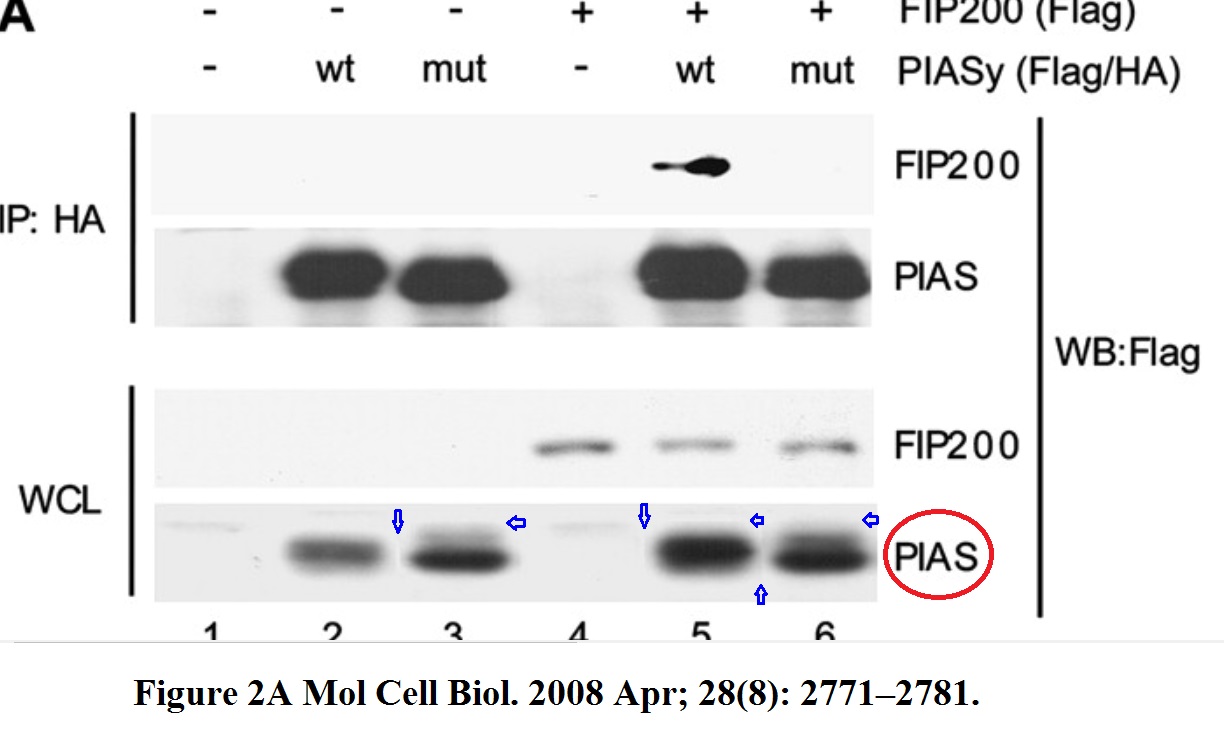
N Martin, K Schwamborn, H Urlaub, B Gan, JL Guan, A Dejean Spatial interplay between PIASy and FIP200 in the regulation of signal transduction and transcriptional activity Molecular and Cellular Biology (2008) doi: 10.1128/mcb.01210-07
Now it might appear like some sloppy but otherwise innocent gel splicing, but it is not. The horizontal splice edges are the giveaway that something more sinister than mere excision of irrelevant gel lanes took place. The PIAS panel in Figure 2A is apparently a collage of various gel bands slapped on in Photoshop, the same thing happened to the PIAS panel of Figure 3C. There however, the bands were not just stuck on randomly: they were precisely arranged in Photoshop on a ladder, to indicate the transgenic vs endogenous PIAS protein. Basically, another PIAS-taking exercise by Anne Dejean and her lab, and yet another reason for the publisher American Society for Microbiology (ASM) to investigate these older works in their journal MCB.
The following is a collaborative paper from the Institut Pasteur, maybe Dejean can check if the problematic figures were made in her lab.
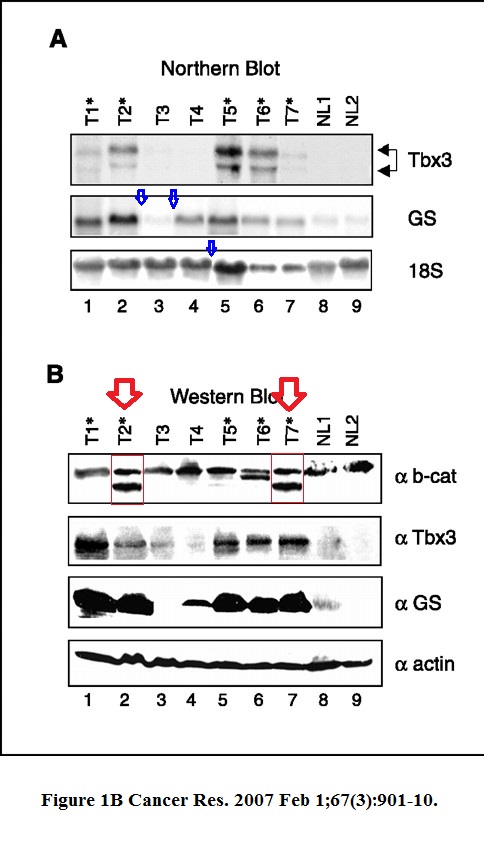
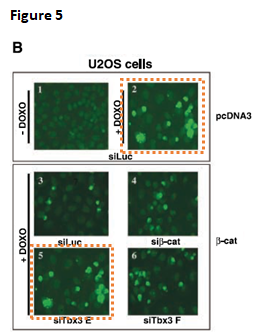
CA Renard, C Labalette, C Armengol, D Cougot, Y Wei, S Cairo, P Pineau, C Neuveut, A De Reyniès, A Dejean, C Perret, MA Buendia Tbx3 is a downstream target of the Wnt/beta-catenin pathway and a critical mediator of beta-catenin survival functions in liver cancer Cancer Research (2007) doi: 10.1158/0008-5472.can-06-2344
Not just some splicing, a b-cat band was most obviously cloned in Figure 1B. Even Nature, where Dejean published quite a lot, was not safe. That journal however replied to Clare Francis and announced to look into the case. On its own, there is just a duplicated SUMO gel:
D Ribet, M Hamon, E Gouin, MA Nahori, F Impens, H Neyret-Kahn, K Gevaert, J Vandekerckhove, A Dejean, P Cossart Listeria monocytogenes impairs SUMOylation for efficient infection Nature (2010) doi: 10.1038/nature08963

But considering what else went on and still goes on in the Dejean and now Dejean-Bischof lab? Well, the worst things ended up strategically in Molecular Cell. We are dealing with real professionals here.
INSERM is already well aware of the PubPeer evidence. Clare Francis also informed Institut Pasteur, and its president Stewart Cole replied to him:
“As is our practice in such matters I have instructed the Committee for Scientific Integrity to conduct an enquiry into the allegations. This will take some time. The PIs concerned have been informed.”
Now I would not get overexcited here. Institut Pasteur previously investigated another affair, relating to their collaboration with the present rector of Karolinska Institutet, and found no data manipulation despite authors admitting a possible gel lane triplication, while the alleged original data did not match.
My prediction: conclusions not affected, original data unfortunately eaten by some goats.
Update 25.05.2021
Bischof has 4 retractions now, 3 with Dejean (Bischof et al MCB 2005, Bischof et al Mol Cell 2006, Ogrunc et al Cell Reports 2016) and one from his postdoc period with Judith Campisi (Bischof et al JBC 2001). Today, I received this email from Christophe d’Enfert, scientific director of Institut Pasteur:
“An enquiry into the alleged case of scientific misconduct by Dr. Oliver Bischof, a CNRS scientist, and Prof. Anne Dejean was launched in the fall of 2019 and recently concluded. Prof. Anne Dejean’s unit at Institut Pasteur is affiliated with Inserm. This enquiry was thus conducted jointly by the scientific integrity officers of the CNRS, Inserm and the Institut Pasteur. Several of the publications linked to the allegation were retracted prior to completion of the enquiry (PMID: 33626350, PMID: 33338405, PMID: 33277404, PMID: 33106371).
The team in charge of the enquiry presented a written report to the heads of the three organizations concerned. The evidence and conclusions indicate that during the preparation of many of the publications O. Bischof, but not A. Dejean, had engaged in fraudulent practices, in violation of the rules of honesty and scientific integrity. After due consultation with the President of the CNRS, Prof Cole, President of the Institut Pasteur, asked O. Bischof to leave the Institut Pasteur without delay. This is why the group of Oliver Bischof is not listed anymore.“

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism, however small it appears to you, will greatly help me with my legal costs.
€5.00






The 2 Oliver Bischof retractions to date give the leadership of the Pasteur Institute (Paris) chance to change the face of science. All the director general of the institute need do is order an institute-wide review of data. Problematic data detected within the institute would likely have ramifications outside the institute leading to a chain-reaction of institution-wide reviews.
LikeLike
3rd retraction for Oliver Bischof, 2nd retraction for Anne Dejean.
2020 retraction notice. https://www.cell.com/molecular-cell/fulltext/S1097-2765(20)30891-1?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS1097276520308911%3Fshowall%3Dtrue
Retraction Notice to: The E3 SUMO Ligase PIASy Is a Regulator of Cellular Senescence and Apoptosis
Oliver Bischof
Klaus Schwamborn
Nadine Martin
Claudio Sustmann
Rudolf Grosschedl
Anne Dejean
Show all authors
DOI:https://doi.org/10.1016/j.molcel.2020.11.049
(Molecular Cell 22, 783–794; June 23, 2006)
We, the authors, were made aware of irregularities in western blots shown in the article. We have further investigated the matter and found that the paper contains multiple examples of incorrect data use in Figures 2D, 4A, 4B, 4E, 6F, and 6H, including duplications and image flipping. All of these figures were assembled by the first author (O.B.). The authors are thus retracting the paper. We sincerely apologize to the scientific community for any inconvenience resulting from the publication and subsequent retraction of this article. All the authors have agreed to this retraction.
LikeLike
A retraction at Molecular Cell means INSERM ordered it, and made Dejean and Bischof request retraction from editor!
LikeLike
4th retraction for Oliver Bischof.
2021 retraction.
RETRACTION| VOLUME 34, ISSUE 8, 108786, FEBRUARY 23, 2021
Retraction notice to: USP1 regulates cellular senescence by controlling genomic integrity
Müge Ogrunc
Ricardo Ivan Martinez-Zamudio
Paul Ben Sadoun
Jean-Marc Lemaitre
Anne Dejean
Oliver Bischof
Show all authors
Open AccessDOI:https://doi.org/10.1016/j.celrep.2021.108786
(Cell Reports 15, 108786-1401–108786-1411; May 17, 2016)
We, the authors, were made aware of irregularities associated in western blots shown in our article. We have further investigated the matter and found that the paper contains multiple examples of incorrect data use and image flipping in four figures, including the vertical flipping and reuse of the panel in Figures 1B and 3D, similar flipping and incorrect blot image in Figure 2C, and incorrect data use in Figure 4A. All of these figures were assembled by the corresponding author (O.B.) who takes full responsibility for the inaccuracies. Under these circumstances, we believe that the most responsible course of action is to retract the paper. We sincerely apologize to the scientific community for any inconvenience resulting from the publication and retraction of this manuscript.
LikeLike
The first author is someone I know personally, we worked in same lab. Good to see Pasteur had Bischof accept all responsibility.
LikeLike
“My prediction: conclusions not affected, original data unfortunately eaten by some goats.”
https://research.pasteur.fr/fr/teams-heads/
Alphabetical list.
David Bikard
Thomsa Bourgeron
Qu’est-ce que ça veut dire?
LikeLike
https://www.cellsenescence.info/
About ICSA
Vice-President,
Oliver Bischof,
Institut Pasteur,
Paris, France
LikeLike
https://www.cellsenescence.info/
About ICSA
President
Manuel Serrano,
Institute for Research
and Biomedicine, Barcelona,
Spain
Retraction.
https://pubpeer.com/publications/A11F4C0AAB8E847EC737DBBB434D49
Multiple examples of image duplication, same data for different experiments.
https://pubpeer.com/publications/76D41452BFD0AE703A3A476E77DFED
Image duplication to the point of nonsense.
https://pubpeer.com/publications/ABB7746FD5493450293B8B1F2BBA61
Deliberate image duplication corrected (middle author).
https://pubpeer.com/publications/0A3B263FB7BA2DEA11CC822875488F
Image duplication.
https://pubpeer.com/publications/7726499A135DF6A597014E16753C36#1
Images very similar.
https://pubpeer.com/publications/A5B124A2F9A5D74EAEBE8223D5669E
Images very similar.
https://pubpeer.com/publications/CC711529FC402CFC74FF61E7C23409
LikeLike
“Multiple examples of image duplication, same data for different experiments.
https://pubpeer.com/publications/76D41452BFD0AE703A3A476E77DFED”
J Immunol. 2006 Sep 1;177(5):3327-36. doi: 10.4049/jimmunol.177.5.3327.
Specific contribution of p19(ARF) to nitric oxide-dependent apoptosis
Miriam Zeini 1, Paqui G Través, Raquel López-Fontal, Cristina Pantoja, Ander Matheu, Manuel Serrano, Lisardo Boscá, Sonsoles Hortelano
Affiliation1Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain.PMID: 16920973 DOI: 10.4049/jimmunol.177.5.3327
Problematic data. Figures 4A and 2. Much more similar after horizontal flips.
LikeLike
Mol Cell Biol. 2000 Mar;20(5):1692-8. doi: 10.1128/MCB.20.5.1692-1698.2000.
Inhibition of IkappaB kinase and IkappaB phosphorylation by 15-deoxy-Delta(12,14)-prostaglandin J(2) in activated murine macrophages
A Castrillo 1, M J Díaz-Guerra, S Hortelano, P Martín-Sanz, L Boscá
Affiliation
1
Instituto de Bioquímica (Centro Mixto CSIC-UCM), Facultad de Farmacia, Universidad Complutense, 28040 Madrid, Spain.
Problematic data figure 2B. Much more similar than expected.
LikeLike
https://cjm2-2020.sciencesconf.org/?forward-action=index&forward-controller=index&lang=en
COMPARATIVE BIOLOGY OF AGING
Due to the current health situation, the conference is postponed (in 2022, date to be determined)
To attend the conferences Jacques Monod, an abstract submission is mandatory,
Registrations request without abstract submission will not be processed, except in specific
cases (i. g. for Publishers : please contact chairperson).
Chairperson: Thomas C.G. Bosch
Christian-Albrechts-University Kiel, Am Botanischen Garten 1-9, 24118 Kiel, Germany
Phone: +49 431 880 4169
Email: tbosch@zoologie.uni-kiel.d
Vice-chairperson: Oliver Bischof
Institut Pasteur, Unité “Organisation Nucléaire et Oncogenèse” – Inserm U993, 28, rue du Dr. Roux, 75724 Paris Cedex 15, France
Phone: +33(0)140613307
Email: oliver.bischof@pasteur.fr
LikeLike
PLoS One. 2010 Jan 14;5(1):e8710. doi: 10.1371/journal.pone.0008710.
Mice lacking thyroid hormone receptor Beta show enhanced apoptosis and delayed liver commitment for proliferation after partial hepatectomy
Raquel López-Fontal 1, Miriam Zeini, Paqui G Través, Mariana Gómez-Ferrería, Ana Aranda, Guillermo T Sáez, Concha Cerdá, Paloma Martín-Sanz, Sonsoles Hortelano, Lisardo Boscá
Affiliation1Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain.
PMID: 20090848 PMCID: PMC2806828 DOI: 10.1371/journal.pone.0008710
Problematic data figures 1A and 2D. Much more similar than expected, although the genotypes are different.
LikeLike
https://www.cellsenescence.info/
About ICSA
President
Manuel Serrano,
Institute for Research
and Biomedicine, Barcelona,
Spain
https://www.ahlresearch.org/manuel-serrano-phd
Manuel Serrano, Ph.D.
ICREA RESEARCH PROFESSOR AT INSTITUT DE RECERCA BIOMÈDICA (IRB BARCELONA)
Manuel Serrano obtained his PhD in 1991 for his research at the Center for Molecular Biology (CSIC/UAM, Madrid)) under the supervision of M. Salas and J.M. Hermoso. From 1992 to 1996 he worked as a Postdoctoral Fellow in the laboratory of D. Beach at the Cold Spring Harbor Laboratory, New York, USA. In 1997, he returned to Spain to start his own research group at the Spanish National Biotechnology Center (CSIC, Madrid). He moved to the Spanish National Cancer Research Center (Madrid) in 2003 to lead the Tumor Suppression Group, where he also served as Director of the Molecular Oncology Program (2012-2017). In May 2017, he relocated to the Institute for Biomedical Research-IRB Barcelona to establish the Cellular Plasticity and Disease Group within the Molecular Medicine Research Program.
What stubborn stain on science this paper is!
Cancer Res. 2005 Mar 15;65(6):2186-92. doi: 10.1158/0008-5472.CAN-04-3047.
p73beta-Mediated apoptosis requires p57kip2 induction and IEX-1 inhibition
https://cancerres.aacrjournals.org/content/65/6/2186.article-info
AUTHOR INFORMATION
Susana Gonzalez1,2, Manuel M. Perez-Perez1, Eva Hernando1, Manuel Serrano2, and Carlos Cordon-Cardo1
1Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York and 2Molecular Oncology Program, Spanish National Cancer Centre, Madrid, Spain
Requests for reprints:
Carlos Cordon-Cardo, Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, NY. Phone: 212-639-7746; Fax: 212-794-3186; E-mail: sgonzalez@cnio.es
PMID: 15781630 DOI: 10.1158/0008-5472.CAN-04-3047
Problematic data at Pubpeer:https://pubpeer.com/publications/ABB7746FD5493450293B8B1F2BBA61
Fresh problematic data figure 3B. Much more similar than expected after horizontal flip.
LikeLike
https://research.pasteur.fr/en/project/transcriptional-gene-silencing-in-cellular-senescence/
If you run your cursor over 01 Feb 2021 this pops up.
LikeLike
“An enquiry into the alleged case of scientific misconduct by Dr. Oliver Bischof, a CNRS scientist, and Prof. Anne Dejean was launched in the fall of 2019 and recently concluded”
Time ended 01 FEB 2021.
Pasteur Institute, Paris should be commended on its swift action!
A shining light in the murky darkness of the biomedical industrial, self-interested complex.
LikeLike
Sanjeev Galande:
Sanjeev used to head the Laboratory of Chromatin Biology and Epigenetics (Sanjeev Galande Lab) at Indian Institute of Science Education and Research, Pune. He is the founder of the Centre of Excellence in Epigenetics at IISER and is known for his studies on higher-order chromatin architecture and how it influences spatiotemporal changes in gene expression.
He is an elected fellow of the Indian National Science Academy and the Indian Academy of Sciences and a recipient of the National Bioscience Award for Career Development of the Department of Biotechnology. He also received the G D Birla award. For his contributions to biological sciences, he was awarded the Shanti Swarup Bhatnagar Prize for Science and Technology, one of the highest science awards in India, in 2010 by the Council of Scientific and Industrial Research.
He sadly has 10 papers of Pubpeer!
Dr. Sanjeev Galande has now joined Shiv Nadar University, Delhi-NCR, as Dean of the School of Natural Sciences and HoD, Life Sciences in June 2021.
LikeLike
“He sadly has 10 papers of Pubpeer!”
It is better that others know, not sad at all.
LikeLike
https://www.liebertpub.com/doi/pdfplus/10.1089/oli.2010.0277?casa_token=BSXaq_4f1PsAAAAA:hUR8zZaQEPYj-9jHHEqpl8HdzNqWHjx_P-oLT7vjtMUCjV4pLq3ZaJxBjhMFC9b7T0OpN7KgDg
OLIGONUCLEOTIDES
Volume 21, Number 2, 2011
ª Mary Ann Liebert, Inc.
DOI: 10.1089/oli.2010.0277
Decoy-DNA Against Special AT-Rich Sequence Binding
Protein 1 Inhibits the Growth and Invasive Ability
of Human Breast Cancer
Asako Yamayoshi,1 Mariko Yasuhara,1 Sanjeev Galande,2 Akio Kobori,1 and Akira Murakami1
1
Department of Biomolecular Engineering, Kyoto Institute of Technology, Matsugasaki, Kyoto, Japan.
2
Centre of Excellence (CoE) in Epigenetics, Indian Institute of Science Education and Research (IISER), Pune, Maharashtra, India.
Sajeev Galande, keep on trucking!
https://pubpeer.com/publications/98AB9B3827A8D62CC1D4A6353E9D11
https://doi.org/10.1089/nat.2010.0277.correx
“The authors of this article entitled Decoy-DNA Against Special AT-Rich Sequence Binding Protein 1 Inhibits the Growth and Invasive Ability of Human Breast Cancer published in Oligonucleotides 2011;21(2):115–121 wish to correct an error.
Two images of decoy-treated MDA-MB-231 cells (Control Decoy (1.7 μM) and SATB1 Decoy (460 nM) in Figure 2C were identical and it was completely an inadvertent error. While submitting a magnified version the authors did not realize the apparent overlap in the images. This error does not affect the inference of the main experiments.
LikeLike
23 Nov 2022 Expression of Concern https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9683845/
For:
PLoS Biol. 2010 Jan 26;8(1):e1000296. doi: 10.1371/journal.pbio.1000296.
Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner
Dimple Notani 1, Kamalvishnu P Gottimukkala, Ranveer S Jayani, Amita S Limaye, Madhujit V Damle, Sameet Mehta, Prabhat Kumar Purbey, Jomon Joseph, Sanjeev Galande
Affiliations collapse
Affiliation
1National Centre for Cell Science, Ganeshkhind, Pune, India.
PMID: 20126258 PMCID: PMC2811152 DOI: 10.1371/journal.pbio.1000296
After this article [1] was published, concerns were raised about similarities and discontinuities between some of the lanes in the western blot images in Figs Figs1,1, 5, S2, and S3, and between some of the lanes in the EMSA autoradiograms in Figure S6.
The corresponding author provided the original underlying images and quantitative data to support several figures in the article (S1 File), but stated that the original data are no longer available for the western blots in Fig 1B and 1D left panel, Figure 4B bottom panel, Figure 4C bottom panels, Figure 7B β-actin panels, Figure S3, Figure S7, Figure S8 and Figure S12 left panel. In some cases, the authors provided replication data to support the figures in question (S2 File).
Overall, the data provided resolved some but not all of the image integrity concerns.
Specifically:
In the left western blot panel in Fig 1D there appear to be vertical discontinuities, and the pixel patterns appear similar between different areas within lane 5. The original underlying data for this panel were not provided and so these issues remain unresolved. The authors stated that lanes were removed during figure preparation, and replication data were provided (S2 File) that support the results reported in the article. In the right western blot panel in Fig 1D lanes 7 and 10 appear similar. The authors confirmed that the background from lane 10 seems to have been duplicated in lane 7. The underlying image provided for this experiment is in S1 File. The backgrounds of lanes 7 and 10 appear different in the underlying blot image (compared to each other and compared to the published figure for lane 7), but underlying data for both lanes indicated negative results. When levels are adjusted to visualise background, there appear to be discontinuities suggestive of image splicing in Figures S2A, S2B, S3, and S6. In Figure S3 the M lanes in the left and centre blots appear similar. The images provided in S1 File to support the left western blot panel of Figure S2A do not appear to match the published figure panel and did not clarify the concern about discontinuities in the published figure. The published figure has a stronger and clearer interaction signal than the underlying images; however, the underlying images- while different from the published figure- show a band in the position that would indicate an interaction, and support the conclusion. For Figures S2B and S6, the original data (S1 File) clarified the concerns. The raw data indicated that there was undeclared splicing in Figure S6 after lane 5 of the CHUK panel. For other panels in these figures the discontinuities observed in the figures were also present in the raw images. For Figure S3, the authors stated that the original data are no longer available and so we were unable to resolve the concerns about the published figure. However, the authors provided replication data (S2 File) that support the results reported in the article. During our assessment of this case, faint bands were noted in the original image (S1 File) for the left western blot panel in Fig 1E that are not evident in the published figure. The corresponding author indicated that they used a lower exposure in the published image, the underlying image for which is no longer available. The data in S1 File are consistent with the conclusion that there is more signal with the PDZ domain than the CD+HD domains, although the difference between lanes in S1 File is not as striking as in the published figure. The authors stand by the conclusions of the study and consider that the additional data provided support all original claims. The corresponding author has provided a revised schematic for Fig 1D below which shows where the two proteins interact and with the relevant amino acids identified.
Overall, PLOS Biology concluded that the article’s main conclusions appear to be supported. However, in light of the unresolved issues and the extent of concerns about the integrity of image data reported in the article, the PLOS Biology Editors issue this Expression of Concern.
Reference
Notani D, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, Mehta S, et al. (2010) Global Regulator SATB1 Recruits β-Catenin and Regulates TH2 Differentiation in Wnt-Dependent Manner. PLoS Biol 8(1): e1000296. doi: 10.1371/journal.pbio.1000296
LikeLike
http://2019.aek-congress.org/oliver-bischof.html
Awards
Prix Scientifique de l’Académie de Médecine Prince Albert 1er, 2007
Prix de cancérologie de la Fondation Simone et Cino del Duca, 2010
Prime d’Excellence CNRS 2012-2017
What a laugh! More fool the committee members who awarded the prizes.
LikeLike
Anne sans Oliver.
https://pubpeer.com/publications/EAB5C9032F714A0B9A64D3A88D9D1A
LikeLike
Oliver Bischof,out from the Pasteur Institute, Paris, in at the Mondor Institute, Paris.
https://eurlive.u-pec.fr/en/pedagogical-team/
LikeLike