Smut Clyde likes his papermills just like he likes his beer. Sure he will go for whatever industrial mass product he can get, but his real connoisseur appreciation goes to little artisan papermills, catering to small select circles of friends and dedicated customers.
Like the little Bo Tang brewery papermill of today’s story, found in China. Somehow the Lawrence Berkeley National Laboratory in California is involved.

Oh what Botangled web we weave
By Smut Clyde
I am not absolutely certain that “Tripartite motif 16 inhibits hepatocellular carcinoma cell migration and invasion” (Li et al 2016) was the first retraction of 2023, but for narrative purposes, let it be so. The retraction was atypically prompt, coming down the track only weeks after PubPeer comments observed that some of its illustrations had stunt-doubles elsewhere in the literature, or identical twins that were separated at birth.

Right Fig 9A from “HOXD9 promotes epithelial–mesenchymal transition and cancer metastasis by ZEB1 regulation in hepatocellular carcinoma” (Lv et al 2015).


Here is a second exhibit: “Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/β-catenin signaling” (Tang et al 2016). Its Western Blot loading controls are versatile:


[right] Fig 2A from “KDM5D inhibit epithelial‐mesenchymal transition of gastric cancer through demethylation in the promoter of Cul4A in male” (Shen et al 2019).
Its xenograft-tumor collections inspire an uncanny sense of déjà vu.
[right] Supplementary Figure S4D from “HOXA7 plays a critical role in metastasis of liver cancer associated with activation of Snail” (Tang et al 2016) [retracted].

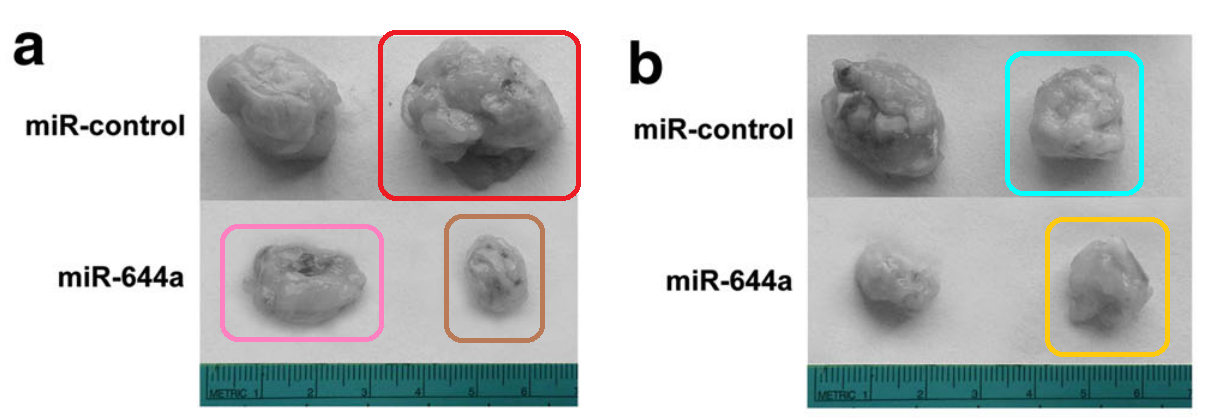
[below] Fig 6a from “MicroRNA-155-3p promotes hepatocellular carcinoma formation by suppressing FBXW7 expression” (Tang et al 2016):
Its cell cultures like to send phase-contrast microphotographs of themselves from different locations, like purloined garden gnomes sending home self-portraits posed in front of tourist destinations around the world.

[right] Fig 3H from “Overexpressed homeobox B9 regulates oncogenic activities by transforming growth factor-β1 in gliomas” (Fang et al 2014)./

In particular, Western Blot bands were shared with “Jumonji AT-rich interactive domain 1B promotes the growth of pancreatic tumors via the phosphatase and tensin homolog/protein kinase B signaling pathway” (Shen et al 2018) and led to the latter’s retraction on 3 January 2023 after an equally brief PubPeer gestation.
All this cries out for an explanation. So here is a spreadsheet instead.



It features one Bo Tang, who has mad networking skills; also enough eloquence to talk international names into accepting coauthorship of fabricated papers. We find him in a series of collaborations with Stephen Tomlinson of the Medical University of South Carolina (Darby Children’s Research Institute)…
And with David Patrick and Yang Yang-Hartwich from Yale School of Medicine…
…Hiroshi Kaneko and Fumio Shimamoto from Hiroshima Shudo University…
[right] Fig 3C from “Expression of USP22 and Survivin is an indicator of malignant behavior in hepatocellular carcinoma” (Tang et al 2015).
…with Fumio Shimamoto and Yun-shan Wang, whose time is divided between Second Hospital of Shandong University and Berkeley Biomedical Data Science Center (Lawrence Berkeley National Laboratory, LBNL)…
[right] Fig 6E from “Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells” (Du et al 2020).

[right] Fig 3A from “Jumonji AT-rich interactive domain 1B overexpression is associated with the development and progression of glioma” (Fang et al 2016).
…and Shijie Cai from Radcliffe Department of Medicine, Oxford University.
[right] Fig 5C from “HOXA7 plays a critical role in metastasis of liver cancer associated with activation of Snail” (Tang et al 2016) [retracted].
[right] Fig 2E from “ABCB5‐ZEB1 Axis Promotes Invasion and Metastasis in Breast Cancer Cells” (Yao et al 2017).
In the earliest papers to attract the attention of the Argus-eyed collective entity of PubPeer (2011-2013), Tang was at the 2nd Affiliated Hospital of Dalian Medical University, working on TCM cancer cures and δ-opioid receptors in the company of his doctoral advisor Liming Wang, helped by the usual scholarly apparatus of falsified Western Blots and flow-cytometry scatterplots / cell-count histograms.*
[right] Fig 5B from “Upregulation of the δ opioid receptor in liver cancer promotes liver cancer progression both in vitro and in vivo” (Tang et al 2013) [retracted]..

Tang was recruited to Affiliated Hospital of Guilin Medical University in time to publish “Silencing the EZH2 gene by RNA interference reverses the drug resistance of human hepatic multidrug-resistant cancer cells to 5-Fu” (Zhang et al 2013).
Fig 2C from “RNAi-mediated EZH2 depletion decreases MDR1 expression and sensitizes multidrug-resistant hepatocellular carcinoma cells to chemotherapy” (Tang et al 2013).
Fig 6A from “Downregulation of δ opioid receptor by RNA interference enhances the sensitivity of BEL/FU drug‑resistant human hepatocellular carcinoma cells to 5‑FU” (Tang et al 2015) [retracted].
Subsequently we find our man at Affiliated Hospital of Xuzhou Medical University, cutting-&-pasting bands for “microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway” (Zhang et al 2018).

[right] Fig 6F from “m6 A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5” (Zhang et al 2019).
 These promotions were accompanied by a cavalcade of colleagues (some of them would be Tang’s own Ph.D students as he rose to professorial status) – each with their own research interests. So Tang’s output broadened to accommodate EZH2 signalling, SIRT / Sirtuin, Ubiquitin, meta-analyses, etc. – pushing back the boundaries of nescience on multiple fronts at once, economically using the same images. This decade-long career through so many universities and hospitals inspired doubts that I had confounded multiple researchers with the same name, and only Dimensions.ai and the continuity of email addresses could convince me otherwise. Note that Tang retains the sheltering aegis of the link to Dalian University, crediting that institution on several more recent papers.
These promotions were accompanied by a cavalcade of colleagues (some of them would be Tang’s own Ph.D students as he rose to professorial status) – each with their own research interests. So Tang’s output broadened to accommodate EZH2 signalling, SIRT / Sirtuin, Ubiquitin, meta-analyses, etc. – pushing back the boundaries of nescience on multiple fronts at once, economically using the same images. This decade-long career through so many universities and hospitals inspired doubts that I had confounded multiple researchers with the same name, and only Dimensions.ai and the continuity of email addresses could convince me otherwise. Note that Tang retains the sheltering aegis of the link to Dalian University, crediting that institution on several more recent papers.
The papermills of my mind
“For as I have often bemoaned in the past, not even the paper-forging industry is free from scruple- and principle-deficient players.” – Smut Clyde
I skipped several academic appointments and connections:
- “Silencing the EZH2 gene by RNA interference reverses the drug resistance of human hepatic multidrug-resistant cancer cells to 5-Fu” involved a secondary affiliation with Dalian Institute of Chemical Physics, Chinese Academy of Sciences.
- After working with Fumio Shimamoto, Tang stayed on at Department of Health Sciences, Hiroshima Shudo University to fabricate “Operative ubiquitin-specific protease 22 deubiquitination confers a more invasive phenotype to cholangiocarcinoma” (Tian et al 2021).
- For “Sirtuin 4 Depletion Promotes Hepatocellular Carcinoma Tumorigenesis Through Regulating Adenosine-Monophosphate–Activated Protein Kinase Alpha/Mammalian Target of Rapamycin Axis in Mice” (Wang et al 2019), he claimed the imprimatur of the School of Pharmacy, Shanghai Jiao Tong University; and Berkeley Biomedical Data Science Center (Lawrence Berkeley National Laboratory). Perhaps the latter affiliation was a ‘visiting scholar’ arrangement, for we have met it before in the context of Yun-Shan Wang. It did not come with an email account for corresponding-author duties.
[right] Fig 8F from “Holliday junction recognition protein promotes pancreatic cancer growth and metastasis via modulation of the MDM2/p53 signaling” (Wang et al 2020)
[right] Fig 4F from “Tripartite motif 16 suppresses breast cancer stem cell properties through regulation of Gli-1 degradation via the ubiquitin-proteasome pathway” (Yao et al 2016).
Of course it is not news to readers that the careers of a research group’s members are sometimes founded on fabricated papers – each paper building on its forerunners and marking progress on a broad research program, but none of them bearing any more connection to reality than the successive volumes of Perry Rhodan.

[right] Fig 3A from “Downregulation of δ opioid receptor by RNA interference enhances the sensitivity of BEL/FU drug-resistant human hepatocellular carcinoma cells to 5-FU” (Tang et al 2016).
But the problems at Dalian Medical University go further; they overlap with problems at Jilin University and others.
The papers are coming from inside the house!
“It feels like half the higher-echelon professors at Jilin University have built their careers on these fairy-tales, with successions of papers itemising the interactions of ADAM10 or GRIM-19. […] if only they had published instead about the Tooth-Fairy circ-RNA and how it targets the Easter-Bunny Pathway…”, – Smut Clyde
Cast your mind back to Tang’s early research on δ opioid receptors, with the flow-cytometry scatterplots I showed earlier. It transpires that other groups were working on the same signalling pathway, at the same time, with no authors in common, but with the same scatterplots. While that recycled suite of cell-count church steeples histograms served to illustrate concurrent work on a different TCM cure for cancer, with no authors in common.
[right] Fig 7 from “Alpinetin suppresses proliferation of human hepatoma cells by the activation of MKK7 and elevates sensitization to cis-diammined dichloridoplatium” (Du et al 2012).
What is unusual is to compare half-a-dozen of these bogus publication trails, from teams at seemingly-unrelated and geographically far-flung institutions, and find them to be coordinated – drawing from a shared pool of images, as if written by a single studio.
All these duplications beckon us down a rabbit-hole of research shenanigans. The rabbit-hole is a spacious warren, with eight branches, marked with colours and letters A to I in the accompanying spreadsheet. All the above only dealt with Branch A.

The spreadsheet also marks ‘intersectional’ entries that share material with other branches, to distinguish them from “intra” and “internal” entries where the duplications are confined to that specific branch, or to that paper.**
Tooth-Fairy-Meets-Easter-Bunny Science of TCM
Smut Clyde follows the dark path of Traditional Chinese Medicine again. What will he find there? As usual, all the big publishers peddling TCM fraud, that’s what.
Please bear with me through the gallery of scratch-assay illustrations that follow, in which the willingness of variously-treated cancer cells to migrate in the manner of amoebae across a glass microscope slide has been measured by gouging a trough through a layer of multiplying cells, and timing how long it takes the neighbours to fill the gap. How well this in vitro ‘healing time’ correlates with cancer invasiveness is a moot point, but fortunately it is not relevant to the popularity of the method. Explanations of ‘migration assays’ and ‘cancer research’ carry little weight if you are caught keying the paintwork on a Mercedes, or so I hear from a friend.

The important point is that the before / after microphotographs of cell migration should be different between experiments, and especially between laboratories. Like finding a trout in the milk-bottle, or feathers on a cat’s face, finding copies in multiple unrelated papers is a legitimate cause to suspect shenanigans. Cell-culture scratches are not the only form of intersectional image being passed around this circle of laboratories like new diseases at a child-care facility. More to come! But first, the other branches!
Branch A provides us with rich veins of laboratory-specific “intra” duplications. Perhaps the images inspired a sense of proprietorial ownership, and their creators were not willing to share them with colleagues at sister institutions. Branch A is largest (56 entries so far) and oldest, with Tang’s 2011 activities. It is not the oldest by much, though; Branch B has entries from 2013.

[right] Fig 6C. from “The role of uPAR in epithelial-mesenchymal transition in small airway epithelium of patients with chronic obstructive pulmonary disease” (Wang et al 2013).
Branch B is a Shandong-specific operation, always involving faculty at Shandong University School of Medicine and / or at hospitals affiliated to Shandong Uni. Forty-six papers (so far) are provided by a number of active contributors; their research priorities include CULL4A signalling (Cullin4A), uPAR and Twist-1, but not to the exclusion of other topics. The most active contributor turns out to be Yun-Shan Wang: Professor at International Biotechnology R&D Center, Shandong University School of Ocean. Already mentioned as coauthor of two of Bo Tang’s papers.
- “Sirtuin 4 Depletion Promotes Hepatocellular Carcinoma Tumorigenesis Through Regulating Adenosine-Monophosphate-Activated Protein Kinase Alpha/Mammalian Target of Rapamycin Axis in Mice” (Wang et al 2019);
- “m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling” (Tang et al 2020).
The scientific sea of miR- and exosome-related knowledge
“Whole cohorts of peer-reviewers have been trained to view all these mannerist stylings as what western blots should look like. […] It will be a challenge to convince them otherwise.” – Smut Clyde.
Branch-B stalwarts Chuanxin Wang and Lutao Du also signed the former paper. The name ‘Yi Zhang’ appears in both branches but may be two different people.
 As noted earlier, Yun-Shan Wang has Lawrence Berkeley National Laboratory connections, of uncertain strength… Wang claims to be Professor at LBNL on his LinkedIn account, while other sources say that he was there on a state-funded postdoc fellowship. At any rate, the connection was robust enough for Wang and his recycled data to join LBNL faculty on a paper about 3rd-hand-smoke tobacco contamination. One Jian-Hua Mao, who ranks at LBNL as Geneticist Senior Scientist, shares coauthorship on (so far) five of Wang’s fraudulent papers.
As noted earlier, Yun-Shan Wang has Lawrence Berkeley National Laboratory connections, of uncertain strength… Wang claims to be Professor at LBNL on his LinkedIn account, while other sources say that he was there on a state-funded postdoc fellowship. At any rate, the connection was robust enough for Wang and his recycled data to join LBNL faculty on a paper about 3rd-hand-smoke tobacco contamination. One Jian-Hua Mao, who ranks at LBNL as Geneticist Senior Scientist, shares coauthorship on (so far) five of Wang’s fraudulent papers.
- “Short-term early exposure to thirdhand cigarette smoke increases lung cancer incidence in mice” (Hang et al 2018).
Fig 3C from Hang et al.; Fig 6 from “Suppression of CUL4A attenuates TGF-β1-induced epithelial-to-mesenchymal transition in breast cancer cells” (Wang et al 2017) [retracted]; Fig 3e from “HOXA7 plays a critical role in metastasis of liver cancer associated with activation of Snail” (Tang et al 2016) [retracted].
Time for some intersectional variety. How about more ID parades of xenograft tumours, shuffled into sundry arrangements?
[right] Fig 8A from Tang et al (2013)..
[right] Fig 1F from “m6 A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5” (Zhang et al 2019).
There is no end to the scenes of murine mass-murder!

Oh dear, another suite.

[right] Fig 4E from “Vascular Endothelial Growth Factor Receptor-1 Activation Promotes Migration and Invasion of Breast Cancer Cells through Epithelial-Mesenchymal Transition” (Ning et al 2013) [retracted].
The Adventures of a Mouse Malignancy Group Portraitist
Smut Clyde was busy with yet another Chinese paper mill. A plastic ruler was deployed.
 In contrast there is no overlap between Branch C and the circles of Bo Tang and Yun-Shan Wang. Xi’an Jiaotong University and its associated hospitals are the unifying theme here, with Yu Yao, Tao Tian and Kejun Nan as frequent authors. This group also produced entries from 2013 and 2014, but the problematic duplications are only with papers from the same branch; the first intersectional entries, indicative of cooperation or at least interaction with another university, were not until 2015.
In contrast there is no overlap between Branch C and the circles of Bo Tang and Yun-Shan Wang. Xi’an Jiaotong University and its associated hospitals are the unifying theme here, with Yu Yao, Tao Tian and Kejun Nan as frequent authors. This group also produced entries from 2013 and 2014, but the problematic duplications are only with papers from the same branch; the first intersectional entries, indicative of cooperation or at least interaction with another university, were not until 2015.
Ash Ra Template
“There is another possibility, though… that C.-C. Sun is the papermill, supplying colleagues around Wuhan with variants of the same paper, with the condition that those donated manuscripts carry self-citation payloads.” – Smut Clyde
The remaining branches are smaller. D and E comprise 11 and nine papers respectively. D is centred on Soochow University. Like A, the principal authors of E are at Dalian Medical University or Liaoning Cancer Hospital and Institute, but there are no authors in common. Now I am bored, and I have run out of things to say about the branches… some may be mere figments of my overactive pattern-spotting module. Instead, consider yet another form of rug that really ties the whole room together. Here are enough intersectional Migration and Invasion panels to rival European history. But with overlaps.

[right] Fig Fig 2c from “miR-223-3p targets FBXW7 to promote epithelial-mesenchymal transition and metastasis in breast cancer” (Wang et al 2022).

Other forms of image are copy-pasted across paper after paper with no concern for authorship or institution – colony-plate proliferation assays, spheroid assays, endlessly versatile Western Blot bands – but the point is made by now.
The Master of the String-of-Sausages
“I am open to the possibility that they both outsourced their Western Blot production to a single, independent Wurst-Meister specialist.” – Dr Smut Clyde, art historian of the Chinese Papermill Renaissance.
Alternative Title: I knew a man Botangles and he danced for you In worn out shoes
Would it be irresponsible to speculate that all these multiple sets of images being handed around from paper to paper all belong to a papermill? An artisanal, bespoke fabrication studio, catering to a select group of clients that sometimes need a publication in a hurry to grease the promotion slipway of a staff member? It would be irresponsible not to speculate!
I’m not saying that the papermillers are lazy, but this happened:
[right] Fig 6 from “Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/β-catenin signaling” (Tang et al 2016).
One possibility is that Bo Tang and his Branch-A colleagues are a source of the papers that decorate this extensive warren, given the skills that they display, the broad spectrum of data forms they recycle across papers. Suspicion also turns to Yun-Shan Wang and his colleagues at Shandong. Shared authorship documents the connection between Tang and Wang. Cooperation among institutions is a good thing, but it may be that the cooperation here is too close.
Tang and Wang are clearly charlatans. What impresses me is the number of fabrications they’ve published as collaborations with international researchers. The LBNL in particular need to ask themselves some serious questions about quality control.
Professor Zhao’s paper mill of fraud
A cancer research professor in China runs a paper mill, sources claim he sells first authorships for a bribe. Problem for his customers: the peer-reviewed papers they pay for, contain fake data.
The usual caveats apply. Some of the entries in my spreadsheet might be papers that were purchased from an independent manufacturer of malarkey, for researchers who buy manuscripts that they can sign are under no contractual obligation to limit themselves to a single supplier (the best predictor of future non-performance is past non-performance). Or they could be in-house productions.**
There are precedents for all this. Readers may remember Guoqiang Zhao, who ascended a ladder of made-up mouse-choir recitals to prominence at Zhengzhou University… meanwhile supplying colleagues with equally fabricated papers as a way of ensuring a regular flow of citations for those papers that he had signed himself. Zhao’s subsequent fall from grace and early retirement turned out to be more cosmetic than real.
Not to forget the nomadic digital calipers and the xenograft choreography from Tianjin Medical University.
Ticket to Tianjin
“So here is a novelty in the annals of fictional research: a nomadic digital caliper. It visited a series of laboratories, accompanied by a backing troupe of mouse-mined xenograft tumours for it to measure” – Smut Clyde
I devoted a spreadsheet to a group of hepatobiliary surgeons at Xi’an Jiaotong University (Shaanxi) but for some reason (most likely cafard or ennui) I only posted about them in passing. Their oeuvre contains the usual impedimenta of Transwell panels, colony plates, excised tumors and repurposed WBs. About half were signed by the group members themselves with the other half bestowed upon colleagues elsewhere at Xi’an Jiaotong University, or further afield. The group are at least responsive, replying to many of the corresponding PubPeer threads. Sometimes they blamed the image errors / manipulation upon poorly-supervised students, and promised that retractions would be expedited; other times they complained that they were the real victims, plagiarised by the clients. 2/10, customer service is abysmal, would not buy from this papermill again.
We began with boring monochrome phase-contrast microphotography, and might as well finish with it. In fact it is a gateway into a garden of bright immunohistochemistry fluorescence:


[right] Fig 8A from “Estrogen promotes stemness and invasiveness of ER-positive breast cancer cells through Gli1 activation” (Sun et al 2014).
Fig 6 from “SET and MYND domain-containing protein 3 is overexpressed in human glioma and contributes to tumorigenicity” (Dai et al 2015) includes all the murine crime scenes with which we began.
Footnotes
* “Reversible and Time-Dependent Inhibition of CYP3A4-Mediated Nifedipine Oxidation by Noscapine” (Jin et al 2011) was published in the predatory Latin American Journal of Pharmacy and can only be accessed by sending money in unmarked bills, so it has not felt the refining fire of PubPeer critique.
** Papers marked with ‘Internal’ and ‘Intra-lab’ could be in-house fakes. Or even genuine errors, spawned by poorly-archived data and an attitude that illustrations of pictorial data are merely serving suggestions that should not be taken literally.

Donate to Smut Clyde!
If you liked Smut Clyde’s work, you can leave here a small tip of 10 NZD (USD 7). Or several of small tips, just increase the amount as you like (2x=NZD 20; 5x=NZD 50). Your donation will go straight to Smut Clyde’s beer fund.
NZ$10.00

























































Oh oooh! Even Berkeley, Yale and Oxford purchase papermill products! What a laugh!!
LikeLiked by 1 person
Finally reading something great after a long wait. Thank you Smut Clyde.
LikeLiked by 1 person
It’s been a while since the last post but I got sidetracked.
LikeLike
So impressive! Time to take it all in.
LikeLiked by 1 person
The huge amount of work put in to untangle and make sense of this is impressive 🤯 I switch off when I read the author list and I see more than one Zhang.
LikeLike
/chuckle when I read “Zhang” as I too have independently come the conclusion there is a genetic predisposition to papermilling and photoshopping in the Zhang family – it’s a turd in the punchbowl of author lists. sorry to all those Zhang’s that may be offended but I can back it up with a quick backend database query on PSIref.
LikeLike