Smut Clyde emerges from yet another dive into the bottomless sea of Chinese paper mill fraud. Look what he caught now, a monster, with over 600 papers spread over dozens of journals!
This time, the paper mill, which Smut Clyde called “Contractor”, also churned out several papers in Frontiers. Which may not sound like surprising or special in any way, but thing is: this Swiss Open Access publisher (owned largely by Springer Nature’s mothership Holtzbrinck) claims to have developed the world’s bestest and reliablest AI tech for quality control of all of science. AIRA, as it is called (for Artificial Intelligence Review Assistant), is aimed to eventually replace the troublesome human reviewers and to screen for EVERYTHING: plagiarism, bad English, conflicts of interests, ethics statements, data completion, scientific value, and of course also image manipulation and duplicated patterns. You can sack all editorial assistants, speed up the acceptance rate to a few hours, and cash in so big your head will spin. Frontiers is so generous that they even offer their AIRA software to paying customers with other scholarly publishers, I mean how cool is that.
In reality, the mighty AI of course doesn’t at all work as advertised, because this is how AI always works. The examples Smut Clyde found in the very recent Frontiers papers include phony western blots (AI generated though!), duplicated images and ridiculously bad hand-drawn flow-cytometry. All fabricated by a giant Chinese paper mill. But it passed both ARIA and the famously “rigorous” peer review at Frontiers with flying colours, so there.

But to be fair, Frontiers is apparently too bloody expensive for that kind of customers from China, what with the APC of ~$3000, so in a way the high paywall might even keep out the trashiest of papermill products, no AI needed. The usual customers of this new mega-mill are the same trash journals we met before, and other outlets by scholarly publishing giants like Elsevier, Springer Nature, Wiley and Taylor & Francis, including Scientific Reports or eBioMedicine, and of course also Blagosklonny’s Cell Cycle, Aging and Oncotarget. Even allegedly respectable society journals were trapped, like FASEB J and (once) Journal of Biological Chemistry.

The whole list is available here, as Google Sheet file.
For our readers behind the Great Chinese Firewall, here a pdf:
Finding a paper not from the mill becomes a cause for celebration
By Smut Clyde
The elevator pitch for J. G. Ballard’s “Report on an Unidentified Space Station” is that it’s a parable by Kafka or Borges, but in space! A crew of astronauts record their hesitant forays into an artefact they have encountered somewhere in the interstellar void, half-obscured in a dust-cloud. At first they down-play its scale. But doors open into corridors, elevators lead to new levels of deserted airport-lounge banality, and the sense of magnitude grows… each time they think they grasp the station in its entirety, it turns out to be a foyer or annex to something much larger. By the story’s end the astronauts are separated and lost in its vastness, but they are not worried about the impossibility of retracing their path to their ship, as the known universe is contained within the artefact they’re surveying.
This is roughly how I feel about the output of the “Contract papermill”. At first sight it was small and specialised, active in the past, characterised by specific formats of fake Western Blots and Flaw Cytometry. But each time I think the whole iceberg has been explored, it proves to be just the tip, as new features show up to extend the net.
The number of identified products has passed 600 (which is my excuse for writing about the mill again so soon), and the question is whether the final tally will remain in the high three figures, or break into the thousands. Either way, I realise that this mill is more productive than any others known so far – even more prolific than the ‘tadpole papermill’; and far from being of largely antiquarian interest, it is as active now as it ever was, churning out flim-flam across a broadening range of topics. Its fabrications comprise a significant portion of the ‘scientific sea’ of miR- and exosome-related ‘knowledge’. Finding a paper not from the mill becomes a cause for celebration.
Those earlier underestimates of the mill’s oeuvre were dominated by three journals: Cellular Physiology & Biochemistry, J. of Cellular Biochemistry and J. of Cellular Physiology (systematic searching may have been involved). Occasional admissions emerged from the putative authors that they had resorted to the services of an outside company for experimental work (and, it may be, for assembling the manuscript and submitting it to a reliably-complaisant journals through a disposable email account). Then the editors and publishers of those journals accepted the evidence that the original peer-reviewing had been suborned or compromised by incompetence, and the travesties they had published were swept away in a tsunami of retractions.

But any sense of making progress was illusory, for additional criteria and cleverer search strategies reveal more infiltrated journals, and another cohort of previously-unrecognised junk papers in the first three.
Here are some highlights, with whole issues becoming papermill vehicles.
- Biomedicine & Pharmacotherapy (28 papers)
- Molecular Therapy – Nucleic Acids (27)
- Bioscience Reports (21)
- Cancer Cell International (21)
- Journal of Cellular and Molecular Medicine (21)
- Oncotarget (18)
- Journal of Biological Chemistry (1)
That last journal was previously celebrated for its hardline, zero-tolerance policy on image shenanigans. Usefully, its on-line archives included each paper’s publication history, with the original version (as shoehorned into the submission template by the authors) as well as the final version after editing, reformatting and pagination.
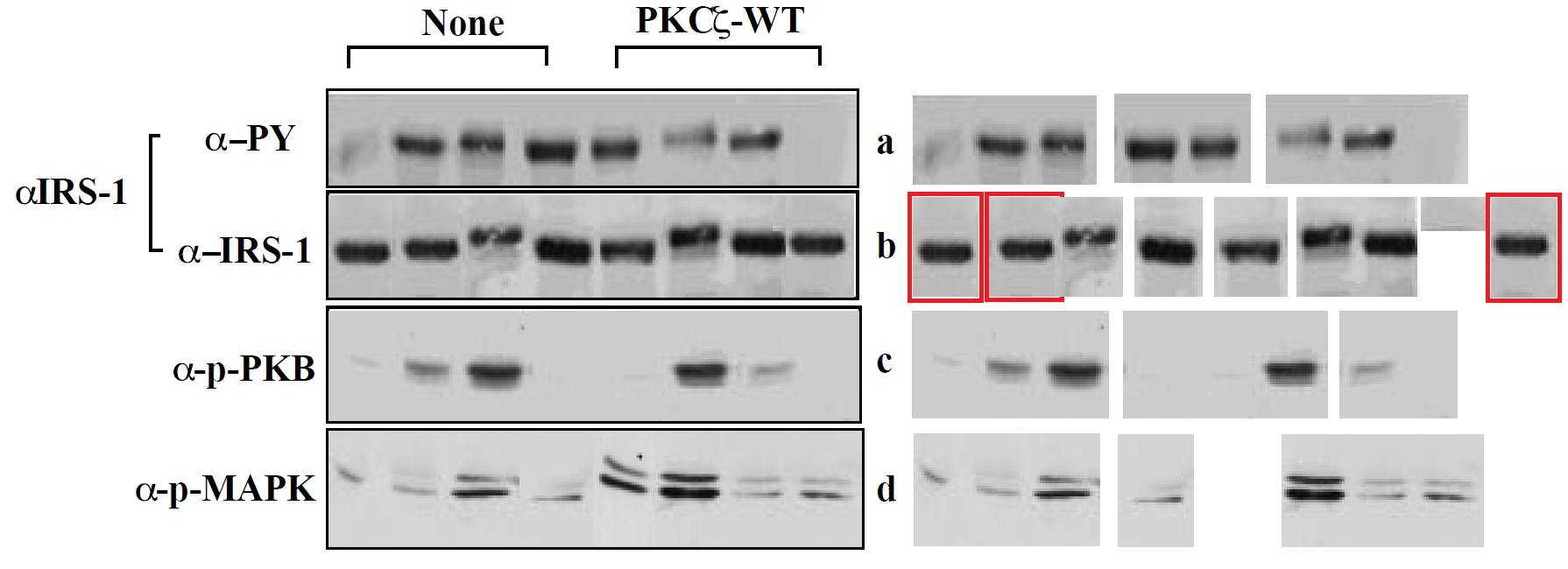
This resulted in surprises for authors who had concocted their Western Blots by building up and superimposing fragments from diverse sources, in more layers than the wallpaper in a student flat, for they had no inkling that their original PDFs allowed interested readers to unpack the embedded images. Hilarity ensued.
JBC‘s reputation for probity contributed to making it a desirable acquisition. To cut the preamble, the new publisher (Elsevier) deleted all those publication-history archives, introduced a Statute of Limitations on fraud, and dispensed with the role of Data Integrity Manager. Even so, there must have been celebrations in the papermill office when they found a Forever Home there for one of their products (Zhao et al 2020).
 The evidence that Zhao et al is problematical is only circumstantial (which is the best kind of evidence). Below at left, behold a montage of Western Blots with a recognisable style: a gamut that ranges from bowties, to spinal columns composed of mutant vertebrae. They are overlaid with a layer of texture, to add verisimilitude (sometimes the background texture repeats). The papers from which these images were garnered all have other forms of malarkey and shenanigans… clockwise from upper left, they are Wang et al (2020); Gao et al (2021); Chen et al (2020); He et al (2021). And at right, the western blots from Zhao et al.
The evidence that Zhao et al is problematical is only circumstantial (which is the best kind of evidence). Below at left, behold a montage of Western Blots with a recognisable style: a gamut that ranges from bowties, to spinal columns composed of mutant vertebrae. They are overlaid with a layer of texture, to add verisimilitude (sometimes the background texture repeats). The papers from which these images were garnered all have other forms of malarkey and shenanigans… clockwise from upper left, they are Wang et al (2020); Gao et al (2021); Chen et al (2020); He et al (2021). And at right, the western blots from Zhao et al.
In isolation, an illustration drawn from a circumscribed visual gamut would not be enough to convict other papers, but it certainly casts a pall of suspicion over them. Notably, we encounter the same style in multiple papers from Aging and in journals from the Frontiers stable (whose proprietors pride themselves of the state-of-the-algorithmic-arts software, AIRA, that screens all submitted manuscripts as a gatekeeper against jiggerypokery). Usually it’s accompanied by bioinformational bafflegab that seems to be part of the template.
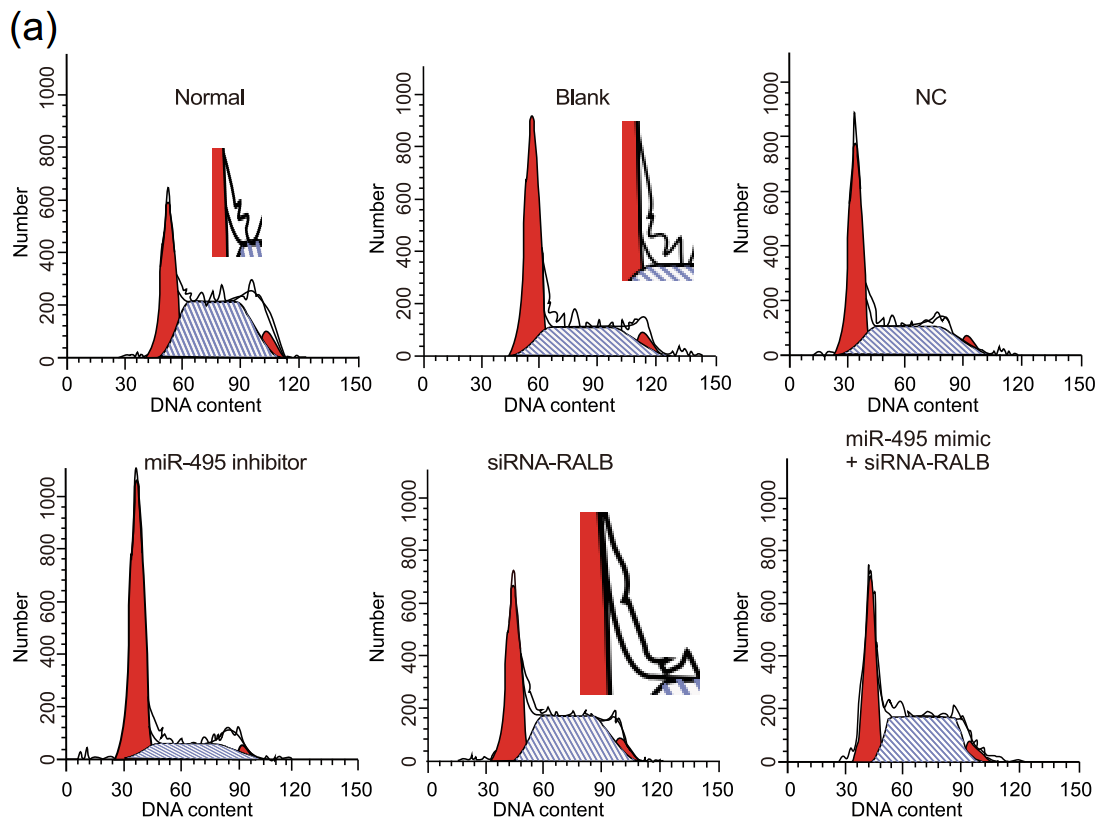
One finds examples of this style of western blots in Lv et al (2020) – retracted for photoshoppery, and for cell-cycle diagrams where the G0/G1 “steeples” appear to have belly fat.
Similar examples appear in this retraction for Gao et al 2020, though they are at the thicker, less regular end of the spectrum. ‘External company’ is here a term of art meaning ‘papermill’. The authors admit that yes, they outsourced the bioinformatic flim-flam and the supposed experiments, but they did collect some basic clinical stats.
The Editors-in-Chief have retracted this article [1] at the request of the authors because concerns have been raised regarding validity of data.
The authors informed the journal that the bioinformatics analysis and all laboratory contents had been performed by an external company. The outsourced laboratory content includes cell culture, FISH, dual luciferase reporter gene assay, RNA pull down assay, RNA immunoprecipitation (RNA IP) assay, cell grouping and transfection, RT qPCR, western blot analysis, chemosensitivity assay and flow cytometry. This was not made clear in this article. The company has not responded to requests from authors to share raw data. Consequently, the data cannot be verified.
The authors stated that they completed the rest of the work, including clinical patient samples immunohistochemistry analysis, statistical analysis and discussion.
All authors agree to this retraction.
In passing, these retractions allow us to identify another papermilling hallmark: flow-cytometry scatterplots that resemble top-heavy pineapples, caught as they topple over.
The topics of western blots and software lead naturally to the scenario in which papermillers train adversarial generative neural nets to synthesise blots de novo, conformant to a style, yet guaranteed to be unique (much as nets can synthesise deepfake faces to grace social-media bots). This worst-case scenario has weighed on the minds of catastrophic doomsayers. I am not so concerned, as implementing it would require investment, whereas the business model of the parasitical-publishing industry is a manifestation of late-stage capitalism – it’s all about maximising the extraction of immediate profit.
More to the point, we are in that CODE NIGHTMARE GREEN dystopiary already, without the intervention of computer learning. Millers are already using existing image-processing software to escape the limitations of ransom-note collage and to generate blot-like images that avoid detection by simple pattern-matching and repetition-spotting.
My art-historical instinct is to take a corpus of all the western blots traced to the present mill and classify them into styles. We do not need AI for that either.
The Five Type of Western Blot
In the best art-taxonomic tradition, I offer five phases. The first three were devoid of backgrounds. The bands remained linear (blobs strung along the horizontal threads of an invisible abacus in a kind of Glass Bead Game), and displayed no progressive development in the morphology of the bands as the proteins migrated across the gels.
- Geometrical dashes, airbrushed into flawlessness, all shiny and chrome for the gates of Valhalla.
- is the ‘ransom-note alphabet‘ notable for the ‘bubble-and-eye’ blobs within its most common loading control, encountered in over 100 papers so far (one of half-a-dozen common recurring loading bands).
- Strings of sausage links. Or if you like, a bakery shop-window display of croissants. These are evidence of fakery, at least in the eyes of the editors of Clinical and Translational Science. Viz. Wang et al (2020):
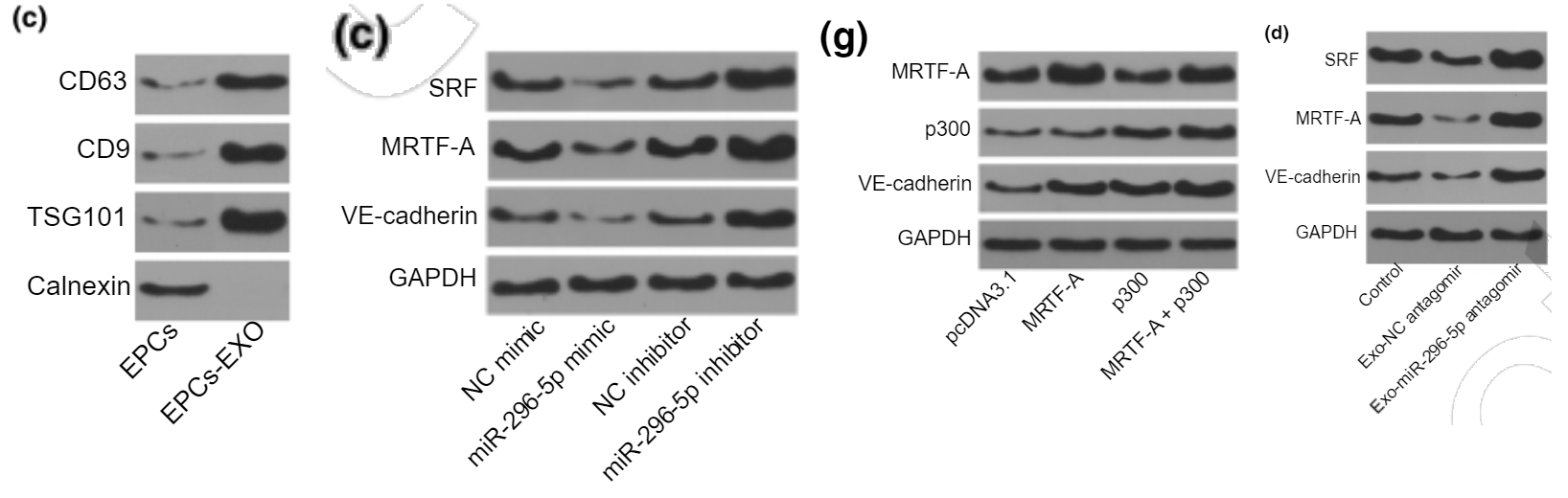
The above article, published online on May 14, 2020 in Wiley Online Library has been retracted by agreement among the journal Editor-in-Chief, Dr. John Wagner, the American Society for Clinical Pharmacology and Therapeutics, and John Wiley & Sons, Inc. The editors had concerns about the validity of the Western blot and qPCR data presented in the article. The authors provided raw data, and software image integrity analysis has confirmed the journal’s suspicions that the Western blots have been inappropriately manipulated. The authors disagree with the decision to retract this manuscript.
4. The bands are high-contrast and blobby, seldom symmetrical, irregular blotches of ink on a grey background (featureless, unless the ink soaks into the paper as filaments). They can be reminiscent of black beetles or grassgrub larvae, with the trailing filaments as legs. If anything, their irregularity is what detracts from their plausibility, as when a band is a doublet (e.g. ERK or JNK or LC3-I and -II), when one might expect the sub-bands to present similar shapes, profiles and horizontal alignment – not following their own lanes. Here is a montage I made earlier, drawing on retracted or problematic papers.


The style featured in Xu et al (2019) – retracted by the Editors of J. Cellular Biochemistry, on account of “allegations raised by a third party” and “several flaws and inconsistencies between results presented and experimental methods described“.
Finally, we have already encountered
5. Here the millers have added the background texture and dialled down the random irregularity to optimise the plausibility of the results. I suspect that it is a development or specialisation of 4, and some papers evince hybrid western blots that combine the background texture with the blobbiness and irregularity of (4). Do we have room here for another blot-related Retraction from Clinical and Translational Science? I think we do. Hi there, Huang et al (2020):
“The above article, published online on February 10, 2020 in Wiley Online Library, has been retracted by mutual agreement among the authors, the journal Editor-in-Chief, Dr. John Wagner, the American Society for Clinical Pharmacology and Therapeutics, and John Wiley & Sons, Inc. The retraction has been issued at the authors’ request. After questions were raised about the validity of the Western blots, the authors expanded the sample size and repeated the experiment. The results could not be confirmed and do not fully support the conclusions made in the article”.

Conversely, examples of the more regular end of Style 5’s spectrum include Li, Zhao & Wang (2021), which the authors withdrew on wholly spurious statistical reasons – perhaps to forestall scrutiny of more serious flaws.

To confirm that the Blobby and Sausage western blots come from the same source, Pang et al (2020) used both.

Now some papermill products exist in two versions with the same authors (or with different authors in one case). The older versions linger in the SSRN pre-print archive as relics of their earlier submission to eBioMedicine, followed by the acceptance elsewhere of alternative versions (or revisions). What commends these cases to our attention is the presence of Style-3 sausage western blots in the first versions, replaced by Style-4 or -5 blotss in the revision (perhaps in response to sausage-related skepticism at the first round of peer-review). For instance, Ou et al (2020) was previously Ou et al (2019).

Xiao et al (2021) was previously Xiao et al (2020). The results remain the same but they summarise different western blots.

To be scrupulously fair, SSRN preprints were not involved in the parallel publication of Shen et al (2019) and Shen et al (2020), which are basically the same paper, though the garbage Figures are in a different sequence. FEBS Press (publishers, with Wiley, of Molecular Oncology) have been appraised of the duplication.


For some reason the Blobby Western Blots of the 2019 version were replaced with something unfamiliar for 2020.

Some other styles are associated with rival papermills so I won’t try to fit them into this time-line. Notably, the ‘tadpole mill’ used its titular ‘zany sardine’ and ‘toboggan’ styles and their variants. The ‘Directly impacting’ mill generates blots in its own form of sausage-links / croissants. An unlabeled atelier encountered at Aging favours a combination of background texture and calligraphy.
Whole cohorts of peer-reviewers have been trained to view all these mannerist stylings as what western blots should look like. Much as they have learned to accept childish crayon scribblings as the acme of flow-cytometry cell-cycle analysis, and to expect apoptosis scatterplots to look like clotted clumps of dots sprayed out with the Photoshop stipple-brush. It will be a challenge to convince them otherwise.
But to get back on track, AI enthusiasts are experts on detecting and deterring academic malfeasance, which is why journals and symposia in the AI / machine-learning community are shining lights of integritude HA HA not really. Based on their own experience of fraud limitation, the enthusiasts regularly propose that screening manuscripts for image recycling or manipulation is easy, boring work that cries out to be handled by image-forensics software or a suitably-trained net. Details are always left as an exercise for the audience.
To be scrupulously fair: packages like Forensically and ImageTwin and Sherloq are becoming helpful tools for the massively-parallel searches for shenanigans within a paper (and for annotating the findings in a way that editors might understand).
I also grudgingly concede that AI might have a place in categorising blotfakes into styles like (4) and (5), if someone (not me!) can train a net on examples until it learns to classify novel examples reliably. Yes, this is an invitation to any enthusiasts out there, to help purge the literature. Of course this would only be a short-term non-solution to the challenge of keeping fake papers out of the literature in the first place, as papermills can switch to new heuristics and continue the arms race. Much as beer is only a short-term solution to the problem of sobriety, but I drink it all the same.



With all that said, above is a sixth style of blotfake, resembling rows of eyebrows, raised in disbelief, with 20 60 examples as of now. In the case of Jia et al (2021), ‘Condylocarpon Amazonicum‘ had insights. I am not 100% convinced that this sixty are the work of the ‘contractor’ mill. They could indicate a new mill (or a new product range from an established mill), encountered at an early stage of the product cycle.
Their other unifying element is an reliance on a particular boilerplate Acknowledgement. The same polite recognition of the role of peer-review has also become one option within the template used for Contractor productions but this could be coincidence.**
We would like to acknowledge the reviewers for their helpful comments on this paper.

I have never plated a boiler myself but I suspect that it’s harder work than it sounds. Anyway, “Boilerplate Acknowledgements” as a part of papermill templates is a topic all in itself. In effect, the real authors sign their work, allowing one to search for their products if any helpful readers feel like adding to the list.
- The authors acknowledge all the reviewers who had given supports for our article
- The authors are grateful to reviewers for critical comments to the manuscript.
- The authors want to show their appreciation to the reviewers for their helpful comments.
- The authors would like to acknowledge the helpful comments on this paper received from the reviewers.
- We acknowledge and appreciate our colleagues for their valuable efforts and comments on this paper
- We express our appreciation to reviewers for all constructive suggestions
- We would also like to thank all participants enrolled in the present study.
- We would like show sincere appreciation to the reviewers for critical comments on this article
- We would like to acknowledge the helpful comments on this paper received from our reviewers.
- We would like to thank our researchers for their hard work and reviewers for their valuable advice
Thanking the reviewers is always courteous, and ingratiating. It’s common enough to thank colleagues (if their contributions were too small to offer them co-authorship or credit them by name). It seems odd, though, to thank the researchers – implying that the researcher was performed by someone else, unless the authors were for some reason acknowledging themselves.
The repetition of these anodyne, time-honoured phrases is not an impeachable offense per se. I could sympathise with authors resorting to previously-published words if they did not trust their command of English enough to thank everyone appropriately in their own phrasing. But in practice that doesn’t seem to be happening. Searching in G**gle Scholar for these Alternative Acknowledgements from the Contractor template turns out to be quite specific in most cases; we are not deluged with false positives (innocent, irrelevant papers, independently crediting people in the same way).* Papermillers are not obliged to use these signatures, and they’ll stop when they realise what they’re doing, but I console myself that they’ll work it out sooner or later, even if it wasn’t spelled out here.
G**gle Scholar returns more hits for these targets than G**gle Search, being better-placed to access and index the Acknowledgement declarations of papers behind a journal paywall.
Rulerpalooza
Still on the general topic of unifying threads in the papermill’s output, a laboratory somewhere in China appears to buy a lot of one specific brand of ruler, for providing scale to xenograft tumours when they excised these from nude mice (the laboratory is also buying a lot of one specific brand of mouse). Then the lab techs treat the rulers almost as harshly as they treat the mice. The end result is eight photographs of a single ruler with a distinctive scratch, spread over seven papers from different authors: Zhou et al (2020); Chang et al (2021) Jia et al (2021); Li et al (2021a); Li et al (2021b); Xin et al (2021); Zhang et al (2021).

In another series of seven photographs and four papers, a (different) ruler is marked by a smudge or a stain: Zhao & Xu (2021); Zhao et al (2021); Gao et al (2021); He et al (2021). Readers may remember some of these from the Bowtie Western Blots montage.

Their repeated manifestations establish that many teams of authors outsource part of their work to a single laboratory. Authors have indeed conceded the point. Even so, this close scrutiny of rulers is obsessive, and I worry about the prospect of becoming a Ruler Truther.
Thank you very much for your comment.
Firstly, I have to apologize for the unreasonable accuse of the authors in the other articles and you for plagiarism.
As for my team, we indeed conducted this experiment outside in a third-party lab, because the condition of my lab is limited to conduct this experiment. This cooperation is legal and not related to any other groups, with no potential conflict of interest.
This lab is located in Jinan, Shandong Province, People’s Republic of China.
The authors in the other three articles may also conduct this experiment in the same external lab.
That Ruler Truther anxiety is such that I content myself with only one more example: Che et al (2020); Liu, Cai & Li (2020); and Yang et al (2021).
Outsourcing experiments is not evidence of data malarkey per se – the accompanying tumours in each of a ruler’s appearances are (usually) different, NO WAIT:

Fig 5a from Zhao & Hu (2019); Fig. 7B from Feng et al (2019)
Evidently the millers have access to enough xenograft tumours that they do not need to recycle, hinting that one of them has a day-job in a laboratory with this specialty. However, their day job does not provide access to Flow Cytometry results (or training), hence all the hand-drawn cell-cycle histograms and apoptosis scatterplots.
The problem, anyway, is that an anonymous external laboratory is also unaccountable, and as seen in umpteen examples, it has strong incentives just to make stuff up. To say nothing from the declarations from the authors that all the work reported in a paper is their own. Analogously, falsely declaring at the Airport Check-in desk that you packed all your bags yourself (when in fact you hired an anonymous contractor to pack them) does not establish that they conceal contrabands or bombs… but airport Security get grumpy all the same, or so I hear from a friend.
As a change from rulers, Leonid asked me to include these hearts, scarred by ischaemia / reperfusion damage from a simulated heart-attack, then extracted from rodents who were no longer using them. Sometimes they are described as belonging to mice (Liu et al 2019) and sometimes as rat-sourced (Wu, Wu & Ni 2019).

They lie on a bench-top surface that could be cracked and perished from age, or alternatively some kind of mass-produced non-slip matting with a standard, non-unique texture. In the latter case, the appearance of the same cracks in images from ostensibly unrelated laboratories would not be a problem, but either way the texture provides a constant scale across the papers. This is a problem, what with some of the hearts being nominally rattish in nature while others are mouse-sourced, and the former should be twice the length and breadth. Fortunately capybaras are too cute and endearing to make convenient laboratory animals so we are spared that additional complexity.


So far not many of these cardiology cases have come to light. They testify to the growing versatility of the millers, who also dabble in osteology, hepatology, atherosclerosis and neurology in addition to oncology.
So here are the key points again:
- The scale of papermill mass-production is even larger than I had guessed, with one single mill placing its manifestations in dozens of journals. Or perhaps I am lumping together multiple mills which copy one another’s distinguishing traits, but I will not lose any sleep about doing them an injustice. Some of the 600-odd papers in the spreadsheet may well contain unforged results, but the mill has contributed to it in some way.
- The millers have a limited supply of valid Flow Cytometry plots (if any). Their clumsy faked substitutes render their work easily recognisable. Fortunately, the journals’ peer-reviewers are equally incognizant of what cell-cycle and apoptosis plots should look like. Other papermills may be more skilled in that department.
- Their products are further characterised by unrealistic Western Blots, drawn from a small repertoire of genres; and by the millers’ use of Acknowledgements as a kind of signature; and by other clues into which we need not go.
Thank you for listening to my SMUTtalk.
Normally I would relieve the bombardment of Western Blots with aesthetic examples of Photoshop enhancement, but space does not permit… readers must browse through the mill’s output by themselves. Instead, here are some bonus western blots with divergent sub-bands, from Ye et al (2020) and Zhang et al (2020).
* * * * * * * * * * * * * * * * * * * *
APPENDICES
 * For many people, the Great Meta-analysis Mishegoss of 2015/15 was their introduction to the concept of papermilling. The pages of journals were being swelled by a wave of inconsequential meta-analysis papers, containing the same stilted wording and the same egregious errors, following the same template and replying on the same suborned peer-review process to ensure that they were accepted, and reaching the same conclusion that
* For many people, the Great Meta-analysis Mishegoss of 2015/15 was their introduction to the concept of papermilling. The pages of journals were being swelled by a wave of inconsequential meta-analysis papers, containing the same stilted wording and the same egregious errors, following the same template and replying on the same suborned peer-review process to ensure that they were accepted, and reaching the same conclusion that Libya Matrix metalloproteinase-7 mRNA and protein expression in gastric carcinoma (or whatever) is a land of contrasts. They may not have been fabricated de novo but “thorough search for relevant studies” was not a priority for the studio while churning out these bagatelles. Then editors caught on belatedly and there was a mighty culling, and the smoke of the burning went up to heaven.
I don’t think anyone has previously commented on the signature Acknowledgements that also unite many of these papers and unite them. Of almost equal importance is the much larger number of inconsequential meta-analyses that survived the great culling despite displaying the same signatures. They’re probably just as bogus, but I suppose no-one pays much attention to them so their extirpation from the literature is not a matter of urgency.
Perhaps the meta-analytical papermillers went on to become the Contractor or Eyebrow atelier, but that would be mere speculation.
** PubPeer contributor Aphilanthops Foxi has commented on several of the threads devoted to ‘Eyebrow’ papers. I am grateful for permission to quote A. Foxi’s following report on those papers. More details have been included in the Eyebrow spreadsheet. Evidently the anonymous scriveners who ghost-wrote the papers are not wise in the ways of micro-RNAs, although they know more about them than I do.
“54 of 55 papers have a single mature miRNA as a main focus.
51 of 55 papers have a “Table 1” with PCR primers. This is usually called “Primer sequence”, occasionally “Primer sequences”. The remaining four articles have the primers in table 2 (3) or 3 (1), likely because of journal requirements for patient characteristics coming first. This is yet more evidence of a common origin.
The mature miRNAs that are the subject of 54 papers can be reverse transcribed only with special techniques and reagents. Subsequent qPCR requires the use of universal primers and, depending on the system, specialized probes. Of the 54 miRNA papers, only 9 report miRNA-specific reverse transcription and/or qPCR methods. Of these 9, 4 report only miRNA methods, suggesting that the longer RNAs are reverse-transcribed and amplified in the same way as miRNAs. Just as miRNAs cannot be amplified by traditional methods, with few exceptions, long transcripts will not be amplified with miRNA-specific methods. Also of note, the primer tables do not list reverse transcription primers (except when these are copied from other papers and confused for reverse primers, a different thing) or probes.
All papers that mention qPCR normalizers in the methods use the same normalizers, U6 and/or GAPDH. Those that do not list normalizers in the methods show primers for U6 and/or GAPDH. For almost half of the papers, the methods suggest that both U6 and GAPDH were used to normalize both miRNA and longer RNA expression. Although U6 is a common miRNA reference transcript for cellular material, and GAPDH for longer transcripts, it is unusual to see them employed so consistently in a group of papers and across so many types of biological samples, including low-abundance materials like EVs, where U6 is not an accepted reference.
All 54 miRNA papers reported on one or at most two transcripts that were putative targets of the mature miRNA. Every paper showed an alignment of the miRNA with its putative target (not unusual in itself). Nine of the published alignments were severely mis-aligned, a jarring mistake that any miRNA biologist would notice immediately. More than 20 of the 54 miRNA papers used a program called “RNA22” to align miRNAs with putative targets. RNA22 is an extremely permissive program and frankly no one uses it anymore. In most cases, the published alignment was non-canonical, with wobble base-pairing, non-canonical seeds, even binding of just a few bases in the middle of the miRNA. These predictions are not stringent. There were 3 main formats of these alignments, with just a few outliers.
Of the 54 miRNA papers, 33 had fatal (non-functional) primer design errors. An additional four or five papers had possible problems, but may have been functional. However, not all primer sequences were checked. For most papers, only the putative miRNA primers were checked. A common theme, with only two or three exceptions, was that the mature miRNA primers were presented as if both forward and reverse primers were specific to the miRNA (incorrect) and not (correctly) as forward (specific) and reverse (universal) primers. A range of errors was detected. miRNA primers were often copied from other papers in which the primers were used for different purposes. For example, cloning primers used to clone a precursor or primary miRNA sequence into a cloning vector were mistaken for mature miRNA primers. Primers for amplifying a precursor sequence were confused for mature miRNA primers. Some primers were for different sequences entirely. Others were for the wrong species and unlikely to function for the target species. Some did not seem to match to any known sequences. In many cases, the forward primer was truly specific for a microRNA, but for the wrong arm (the wrong mature sequence, of two possible arms produced from a precursor). This meant that many studies, assuming the measurements were done at all, were based on incorrectly interpreted preliminary data. Yet somehow it all worked out in the end, with large, expensive animal studies giving positive results.
15 of the 55 articles included studies of extracellular vesicles (EVs), usually incorrectly classified as “exosomes” without demonstration of endosomal origin. Another one had a diagram on exosomes, but without any corresponding data or text…they must have forgotten to buy the “exosome package!” With one exception, the studies used precipitation or ultracentrifugation to obtain EVs. Precipitation is extremely non-specific, and UC is only slightly better. These methods are now mostly deprecated in the field unless done with care and for specific reasons. Several methods sections betrayed a lack of familiarity with the steps and purposes of the different protocols. 5 showed uninterpretable electron micrographs. Most had an EV characterization pattern of “EM, NTA, Western blot”, but for most, only one sample was shown. Western blots all included a distinct CD63 band, even though CD63 almost uniformly presents as a large smear. Only 2 studies showed “negative” markers. Studies that labeled EVs used non-specific, promiscuous dyes that label everything and transfer easily. The EV methods and reporting were poor but not unremarkably so. As for the miRNA studies, I suspect that little or no actual EV research was done here.
Overall, there was little evidence that anyone involved in assembling or reviewing these papers was well-versed in microRNA biology and quantitation, or in EV studies.“

Donate to Smut Clyde!
If you liked Smut Clyde’s work, you can leave here a small tip of 10 NZD (USD 7). Or several of small tips, just increase the amount as you like (2x=NZD 20; 5x=NZD 50). Your donation will go straight to Smut Clyde’s beer fund.
NZ$10.00




























Thank you, Smut Clyde and Leonid, for taking a look at the mass of fraudulent exosome papers that keep getting published.
As someone working in the exosome field, every day I get notification of yet another miRNA-in-exosomes paper that is completely incompetent at best and highly suspect at worst.
LikeLiked by 1 person
Message from IUBMB Life Editor-in-Chief Stathis Gonos:
“Dear Mr Schneider,
Thank you for contacting us. Kindly note that IUBMB Life has a totally new Editorial teal since January 2021. As a new Editor-in-Chief soon after I took over I have realized the problem you refer to and to this end I have implemented the following actions:
Given all these measures the number of submissions has dropped to about 60% in comparison to 2020 submissions and moreover the number of currently accepted articles is less than 10%.”
Gonos also informed me that the previous EiC who published all that paper mill fraud, was Angelo Azzi.
LikeLiked by 1 person
No surprise each bogus manuscript gets the green light !
LikeLiked by 1 person
Pingback: Trionfi di ieri e di oggi – ocasapiens
Human Gene Therapy (published by Liebert) issued this retraction statement I wholly approve of.
https://www.liebertpub.com/doi/10.1089/hum.2018.212
LikeLiked by 1 person
Pingback: The Rise of the Papermills – For Better Science
Pingback: The Master of the String-of-Sausages – For Better Science
Pingback: The papermills of my mind – For Better Science