When Smut Clyde started poking and prodding the Chinese paper mill industry in early 2020, little did he know how high up the fraud will go. The initial assumption that it was just the occasional little doctors in provincial hospitals who would buy fake papers from inept small-time forgers evolved into a revelation that fake science is a Chinese national industry, operated by entire universities. There seem to be central paper mill departments manned by university employees, tasked with supplying fake papers with fake data to be published in real journals with real Chinese university professors as authors.
China, where state and private property are intertwined and all industry is controlled by the Communist Party, has successfully applied the same method to industrialise research fraud. The western publishers, including Elsevier, probably do not really mind, because for every retracted mill product thousands more get published, a never-ending stream of cash, all to the glory of China’s gigantic output on research papers. Now Smut Clyde uncovers yet another university-run paper mill, this one at the Jilin University.

Now, you may think, pah, probably some dusty village college, who cares. But Jilin University is quite fancy actually, here from Wikipedia:
“Jilin University (JLU; simplified Chinese: 吉林大学; traditional Chinese: 吉林大學; pinyin: Jílín Dàxué; often abbreviated JLU or 吉大) located in Changchun, founded in 1946, is a leading national research university under the direct jurisdiction of China’s Ministry of Education.[4] It is a Chinese Ministry of Education Class A Double First Class University.[5] It is strongly supported by state key projects such as Project 985, Project 211 and Project 2011. Jilin University is consistently ranked as one of the most prestigious universities in China,[6][7] […] The CWTS Leiden Ranking table 2020 ranked Jilin University at 14th in the world based on their publications for the time period 2015–2018.[25]“
So yes, the massive paper mill operation under the guise of a university is “under the direct jurisdiction of China’s Ministry of Education“, who surely will take any complaints about the Jilin fraud industry very seriously.
We now even, for the first time ever, obtained evidence that these Jilin University “scientists” used their papermill forgeries to apply for promotions. For example, Dongxu Zhao‘s successful application proudly listed his paper mill fabrications, like this and this.
Here another such Jilin scholar, Xiao Luo, who also earned a promotion to the rank of professor by listing his fake paper mill “research”. A retraction for fraud was not an impediment, maybe Luo was rewarded for discovering a breast cancer pandemic in men:

Also Zhen Tian got promoted despite a retraction and other paper mill fraud he published:

Here are others, listed:
| Selected researchers in China-Japan Union Hospital of Jilin University who used suspected papermill products seeking promotion. May, 2021 | ||
| Name | # of papers on Pubpeer | Promotion granted |
| Dongxu Zhao (赵东旭) | 6 | Yes |
| Rangjuan Cao (曹让娟) | 1 | Yes |
| Xiao Luo (罗晓) | 5, including 1 retraction by the journal | Yes |
| Xueman Lyu (吕雪漫) | 2 | Yes |
| Zhen Tian (田真) | 2, including 1 retraction due to plagiarism | Yes |
| Jing Li (李晶) | 1 | No |
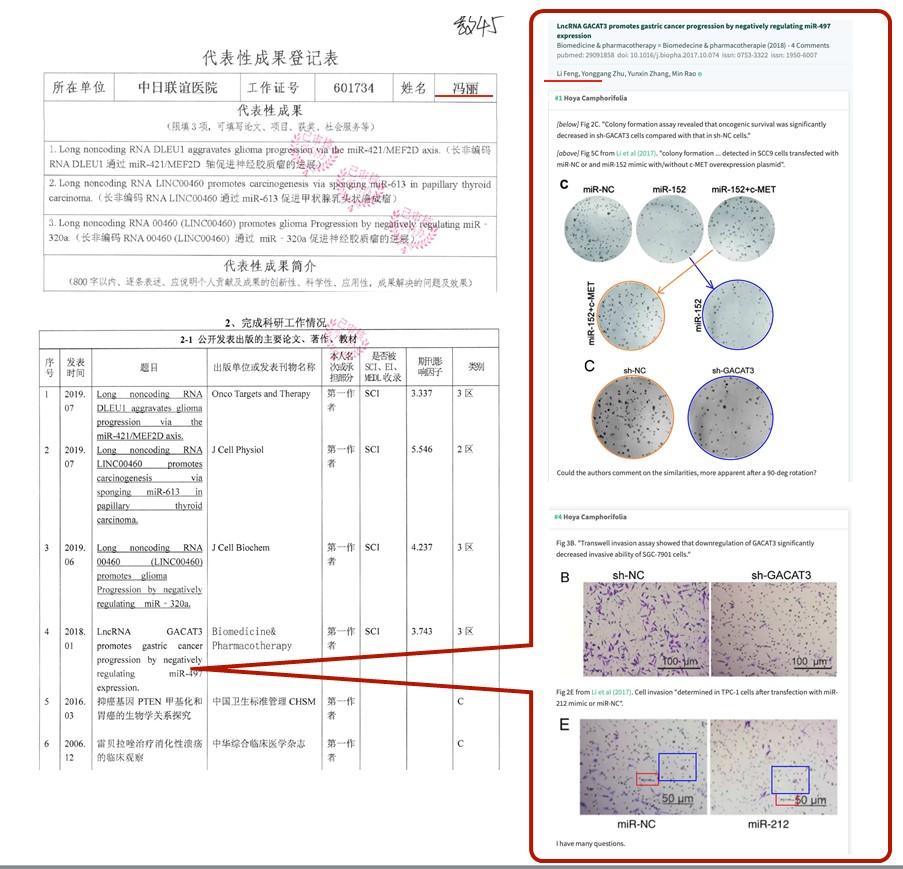
| Li Feng (冯丽) | 1 | Yes |
Think of all the many honest Chinese scientists in Jilin and elsewhere who will not get any promotions or grants, or even lose their jobs, all because they refused to commit fraud. Not just in China: much of our western research, case at hand into non-conding RNAs, is built on referencing such fake Chinese studies. Our fraudsters prosper citing Chinese paper mill fraud to support their own, while our honest scientists are trampled underfoot. Our western editors sit on the editorial boards of these allegedly peer-reviewed journals owned by allegedly serious western scholarly publishers, rubber-stamping fake paper mill products literally on the daily basis. Research fraud is a globalised industry.
Smut Clyde’s Google Sheets list of Jilin’s paper mill activities, with almost 150 papers, is here, for our Chinese readers behind the Great Firewall, a pdf below.
The papers are coming from inside the house! Part 2
By Smut Clyde
So it turns out that the communal laboratories and the horizontal transfer of experimental results among research groups are not confined to Central South University. Data want to be free!
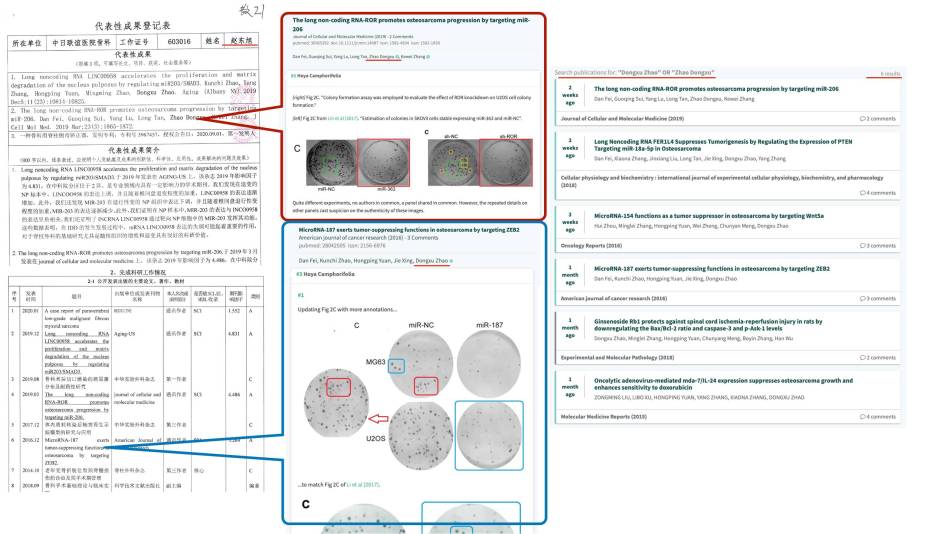
Dear Hoya Camphorifolia,
Thank you very much your comment. We performed these experiments in a public lab that many doctors conduct their studies there. All images were stored in the same public computer, which caused some images misuse. We will thoroughly examine the raw data and lab records of this paper, and upload correct figures to journal and pubpeer. Thank you very much again pointing out this question.
Best regards
Xiaomeng Zhang
April, 16,2021
Dear Hoya Camphorifolia, Thank you for your comment, we will examine orignal data and images, and reply your concern soon.
Best regards
Yarong Li
April,19,2021
Dear Hoya Camphorifolia,
Thank you for your comment, we will examine orignal data and figure to reply your about this question.
Best regards
Huang Wu
I should explain from the beginning. Which is to say, before getting down to the interesting details, this is my chance to indulge in all those vague generalities that no-one would bother reading if they came at the end of a post.

Fig 3A from Jin et al (2015) [19]; Fig 5D from Li et al (2017b) [40]
The systematic invention de novo of an entire research program is a familiar spectacle from US and European science: that is, the manufacture of a series of papers, progressively characterising the cancer-causative or -curing qualities of gene X or protein Y or signalling pathway Z. It is unusual, though, to encounter multiple parallel strands of fabrication within a single institution, using the same building blocks, in a pattern that transcends departments and research groups. Hence my interest in the present oeuvre of 30+ 50+ 80+ over 90 papers.
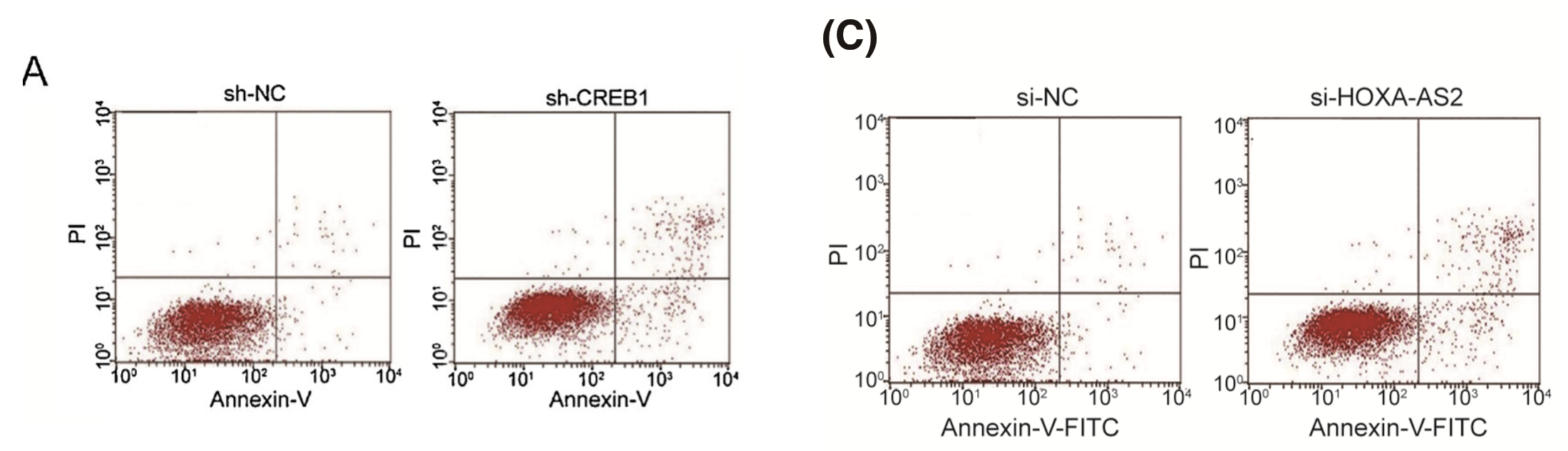
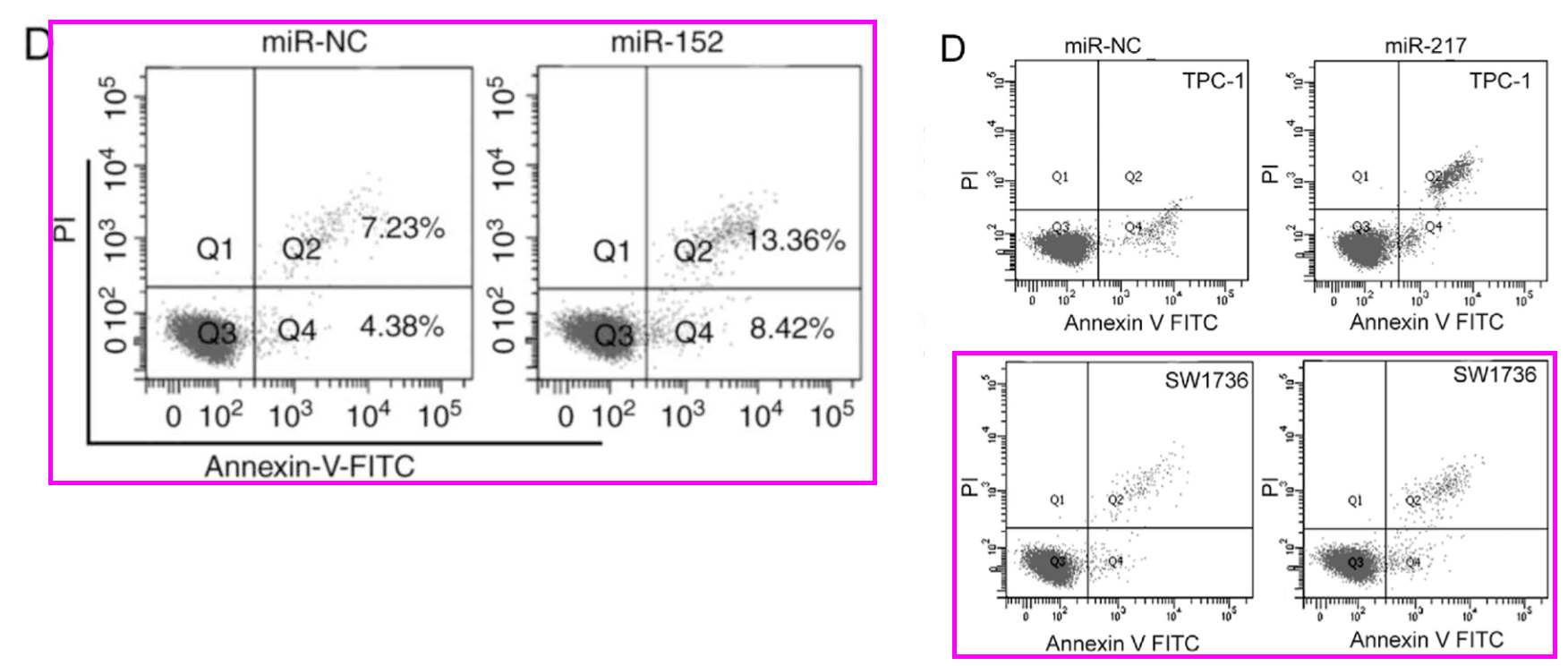
Fig 2D from Guo et al (2018)[42]; Fig 2D from Lin et al (2017) [37]
The authors span the administrative structure of Jilin University and its diadem of associated hospitals, while also spanning the gamut of research directions (the salient word there is “research”, as these authors are career academics with lovingly curated CVs, not clinicians who just need one paper). However, they all dip into a common pool of versatile Western Blot bands to cut up and rearrange in collage. Arrays of xenograft tumors are shuffled into new identity parades in papers with no authors in common. Colony-formation Petri plates are equally Protean, changing labels and acquiring (or losing) the dots of cancer cells proliferating (or not) after some treatment. It is as if someone in central administration realised that to have each section duplicating the infrastructure of forgery was an inefficient waste of resources, so they established a central Falsification Bureau, to provide any department with junk papers and bogus illustrations tailored to their supposed specialties.
The oeuvre appeared mainly in Spandidos journals… branding out into American Journal of Cancer Research and American Journal of Translational Research and International Journal of Clinical & Experimental Pathology from e-Century; and to Tumor Biology from Sage (previously a Springer asset, transferred between publishers in 2016). Smaller metastases exist in Oncology Research and OncoTarget and even Elsevier. The inevitable spreadsheet is here.

Meta-analysts as mudlarks
Now there exists a whole tier of journals favoured by papermills (or symbiotic with them). They are woven into a kind of demi-monde literature or parallel universe by a high rate of mutual citation (with papers from Journal A citing the imaginary results of papers in Journal B to make their own results seem less imaginary), but they penetrate into the real research mainstream when their titles and abstracts are laundered through garbage-in / garbage-out literature reviews and meta-analyses. That is a topic for another day, and here I only want to point out how well the present collection samples this tier. You must wait for some other time to comparisons between literature reviewing and the mudlarks of Victorian London.
It feels like half the higher-echelon professors at Jilin University have built their careers on these fairy-tales, with successions of papers itemising the interactions of ADAM10 or GRIM-19. This is nice for their students / co-authors who thereby ascend the first steps of their own careers, but if only they had published instead about the Tooth-Fairy circ-RNA and how it targets the Easter-Bunny Pathway, there would be less collateral damage to other researchers who take this jibber-jabber seriously.
If this is a Jilin-specific centrally-sanctioned forgery factory, as the evidence implies, it may be that we need a better term than ‘papermill’. ‘Studio’, perhaps. The Central South University collaboration with Spandidos journals is similar but less specific.
To sustain the fiction that the numbers arrayed in tables or plotted as bar-graphs are the results of actual measurements and were not just pulled from the air, papers need some combination of Western Blots; colony formation plates; scratch-heal migration assay and Transwell invasion assay microphotographs; flow-cytometry histograms or FACS scatterplots. So an early eruption of creativity in 2014 equipped this (hypothesised) studio with a storeroom full of the backdrops and stage-props and stage machinery of Theatrical Scientification, to be recycled in productions and performances through 2015 to 2018.
For some tangential reason, a passage from ‘The Reproductive System’ comes to mind:
This literature is full of little anapests.
A sufficiently obsessive reader might recognise the repeated images of xenograft tumors in this collection of papers, and organise them into non-overlapping suites or kunstkammern. You might compare them to shucked oysters, or mutant Gummibären, or wads of used chewing-gum, or diseased brains (depending on your imagination and level of hunger).

Not actually a pavane
In the following montage, tumors dance an stately pavane upon a sheet of blue fabric while transporting patches of adjacent backgrounds around themselves, thanks to the wonders of Appearance-Enhancing Software.
In the left-hand column are Fig 7A from Zhang et al (2014) [3] (“Tumor weight in treated and untreated mice for 21 days“); Fig 6B from Wang et al (2017) [29] (“Representative images of tumors from the TPC-1/miR-497 and TPC-1/miR-NC groups“); Fig 7A from Li et al (2016) [24] (“Photographs of tumor tissue from nude mice 35 days after inoculation“); Fig 6A from Liu et al (2015) [9] (“Graphs of the tumor tissue from the different groups“); and Fig 7A from Zhang et al (2015) [5] (“Images of tumor tissue with different groups collected after sacrifice at day 28“). The right-hand column consists of Fig 5A from Yang et al (2015) [11] (“Images of tumor tissue from different treatments collected after sacrifice at day 22“); Fig 6A from Qu et al (2015) [8] (“Images of tumor tissue with different plasmid treatments collected after sacrifice at day 21“); and Fig 7A from Tian et al (2015) [6] (“Images of tumor tissues in the different mouse groups collected after sacrifice at day 28“).

Alert observers will notice the absence of any consistent lighting in these identity parades, with shadows and highlights differing from one wad to the next, indicating (as if the discontinuities of background were not enough) that their encounters occurred purely in the digital domain.
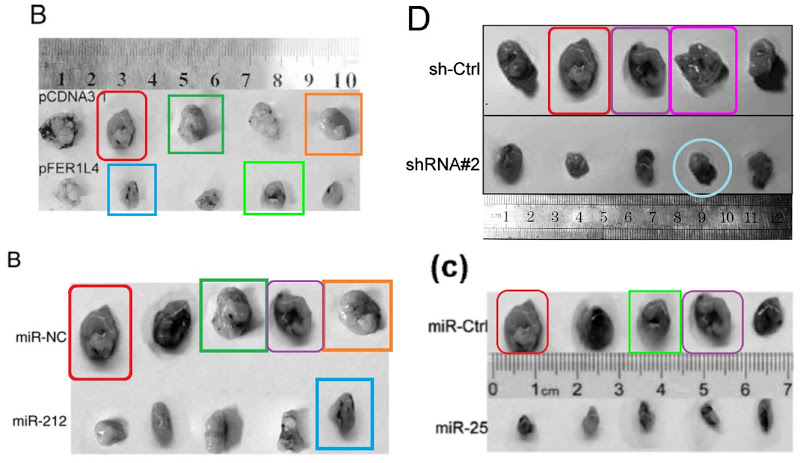
Two more montages are brought together by a ruler that was invited into both dances (or perhaps it’s a Zygmotic Pipette). One consists of Fig 6B from Li et al (2018) [43] (“Tumor tissues were imaged after the mice were sacrificed“); Fig 6B from Yu et al (2016) [32]; Fig 2D inset, from Xia, Li & Lv (2016) [28] (“The mice were sacrificed at 30 days after inoculation, and the tumors were weighed“). The other contains (clockwise from top left) Fig 3B from Fei et al (2018) [45] (“Shown are images of xenograft tumors at 28 days post injection“); Fig 2D from Liu et al (2019) [46] (“Photographs of xenograft tumors“); Fig 7C from Chen et al (2017) [36] (“Photographs of xenograft tumors isolated from miR-25 mimic and miR-Ctrl injected mice“); and Fig 6B from Li et al (2017) [39] (“Photographs of xenograft tumors“).
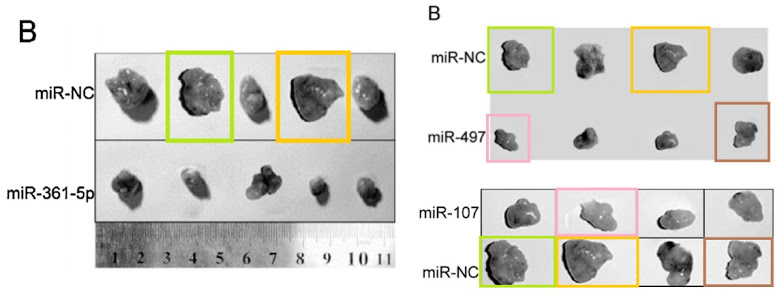
If the (different) ruler is to be trusted in the next set of confections, the tumors change in size alarmingly from one ID parade to the next. They also rotate through 90° or 180° flips. Clockwise from upper left: Fig 7A from Liu et al (2014) [4] (“Images of tumor tissue with different plasmid treatments collected after sacrifice at day 35“); Fig 7A from Zhang et al (2015) [7] (“Images of the tumor tissues from the different groups“); Fig 8B from Li et al (2016) [21] (“Graphs of the tumor tissue”); Fig 7A from Pang et al (2016) [22]; Fig 7 from Wu et al (2015) [20]; Fig 5A from Li et al (2015) [13] (“Images of tumor tissue with different group collected after sacrifice at day 21“); and Fig 5B from Luan et al (2015) [23] (“Photographs of tumor tissue with different group at day 30“).
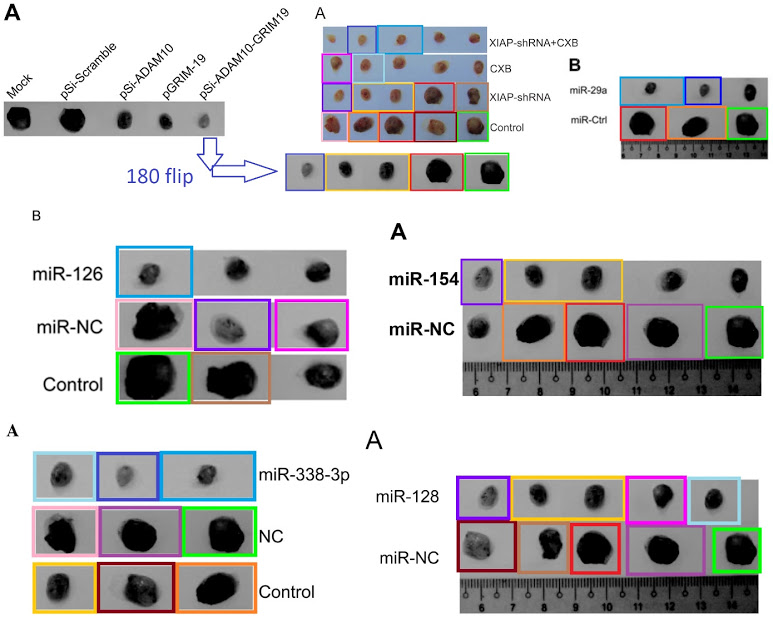
PubPeer contributor “Dendrodoa Grossularia” had already flagged [4] for plagiarising Transwell images from a blameless 2013 paper, which might provide a clue as to the provenance of any material in this whole genre that is not simply repeated with rearrangement and manipulation.
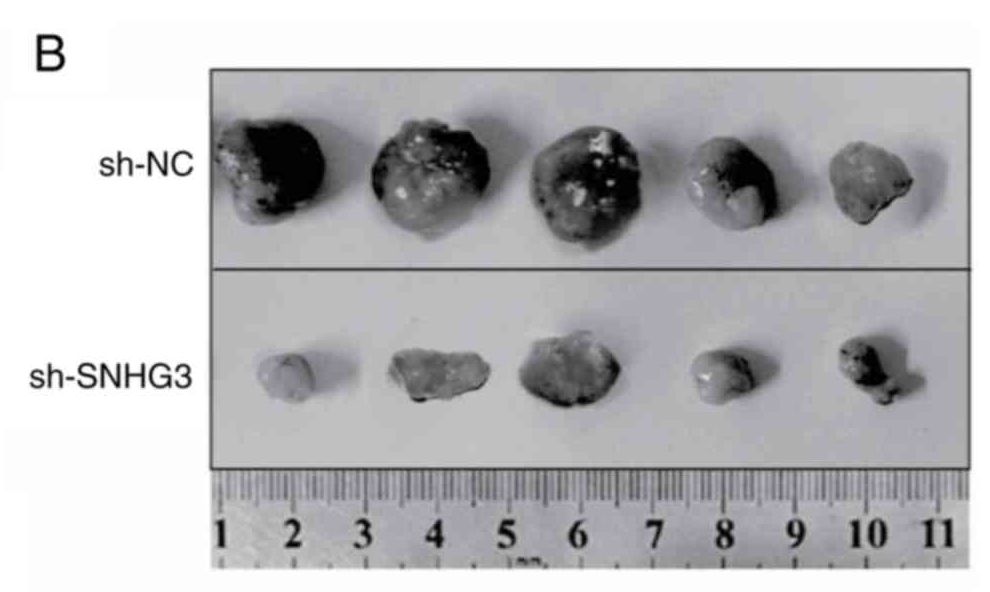
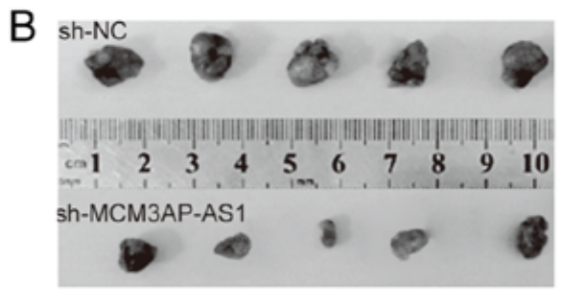
I could flagellate the dead horse further but you get the point: the main item of laboratory equipment for biomed faculty at Jilin University is Photoshop. And rulers Zymotic Pipettes. One particular pipette became the star of Fig 6B from Zhang et al (2020) [50] (“Representative image of isolated tumors from nude mice in the sh-NC and sh-SNHG3 groups”); Fig 7B from Fei et al (2020) [49] (“Representative images of formed tumors that were subcutaneously injected with U2OS cells stable transfected with sh-AFAP1-AS1 and sh-NC plasmid”); …
… Fig 6B from Wen et al (2020) [50] (“Tumor images was captured at the end of the experiments“); and Fig 7B of Zhang et al (2019) [48] (“Representative images of xenograft tumours“).
By the same token, Western Blots in this oeuvre are dominated by six suites, rising and falling in popularity over the period of interest. The robust black blobs of Group-1 show up in 14 papers. They require a palette of coloured boxes to unravel their permutations, and the result is a chromatic catastrophe straight from Edward Tufte’s worst nightmare. These annotated versions should come with a Migraine Trigger Alert for readers who spent last night drinking too much tequila. Would it help if I turned them into strobing animated GIFs?


Fig 1B from Gao et al (2015) [10]; Fig 5C, Li et al (2014) [2]
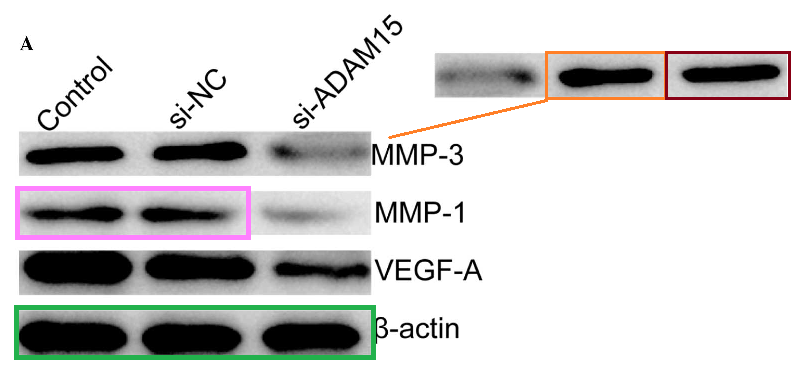
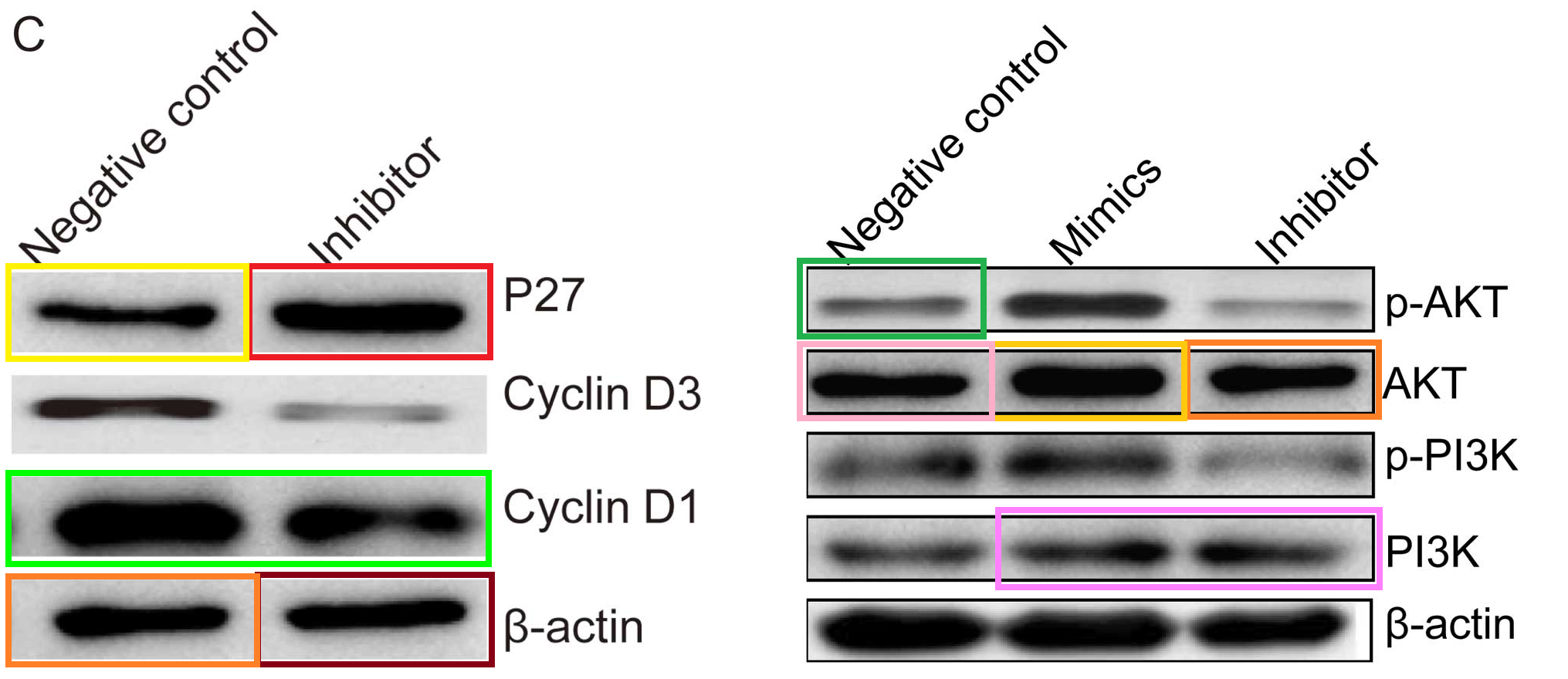
Fig 5A from [10]; Figs 3C and 7 from Liu et al (2014) [1]
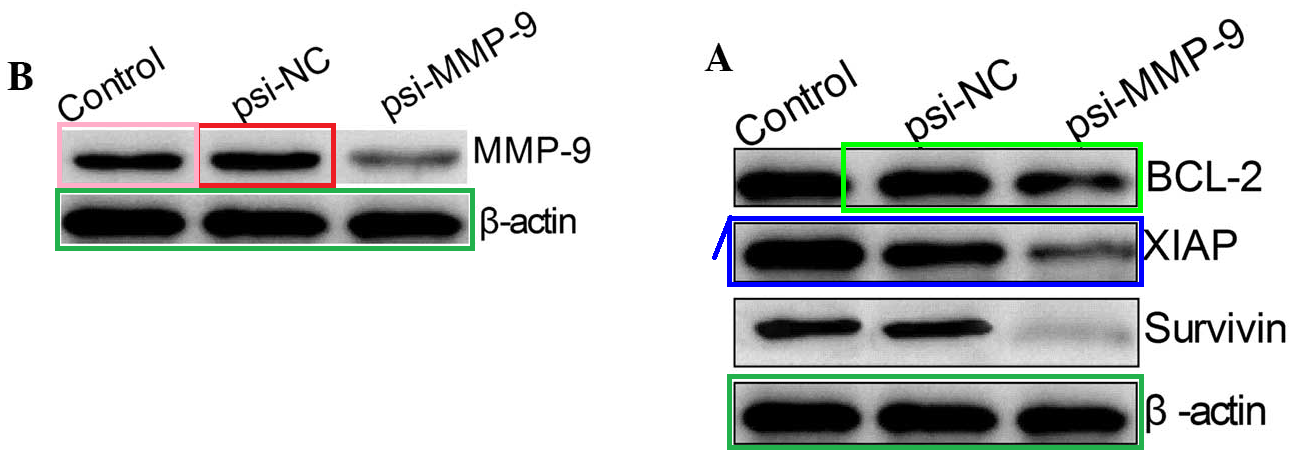

Figs 1B, 3A from Guo et al (2015) [12]; Fig 4 from Li et al (2015) [17]
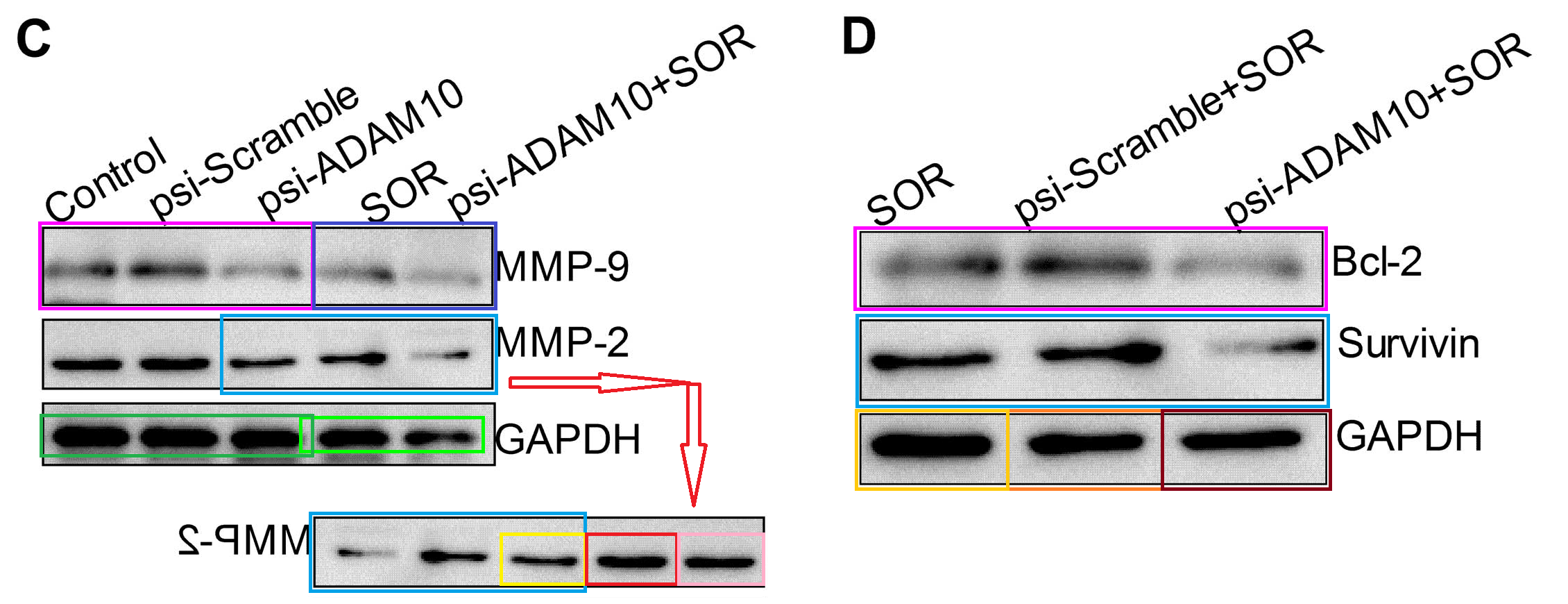

Figs 4C, 5D from [3]; Figs 2C and 5 from [4]
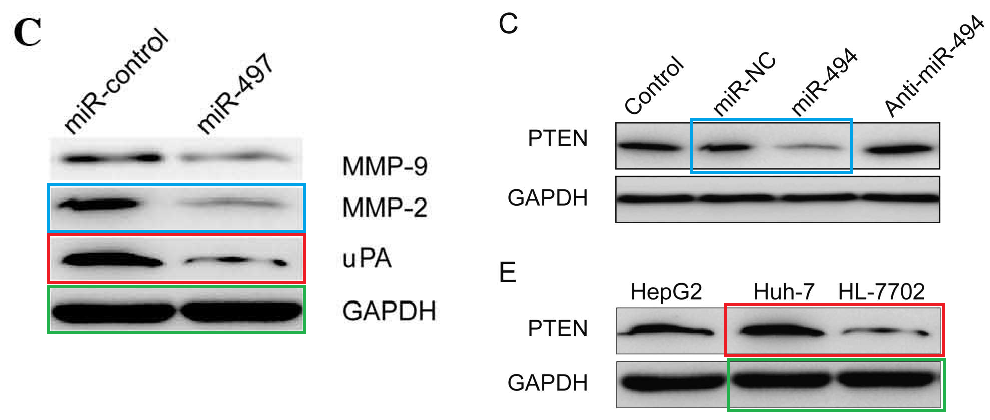
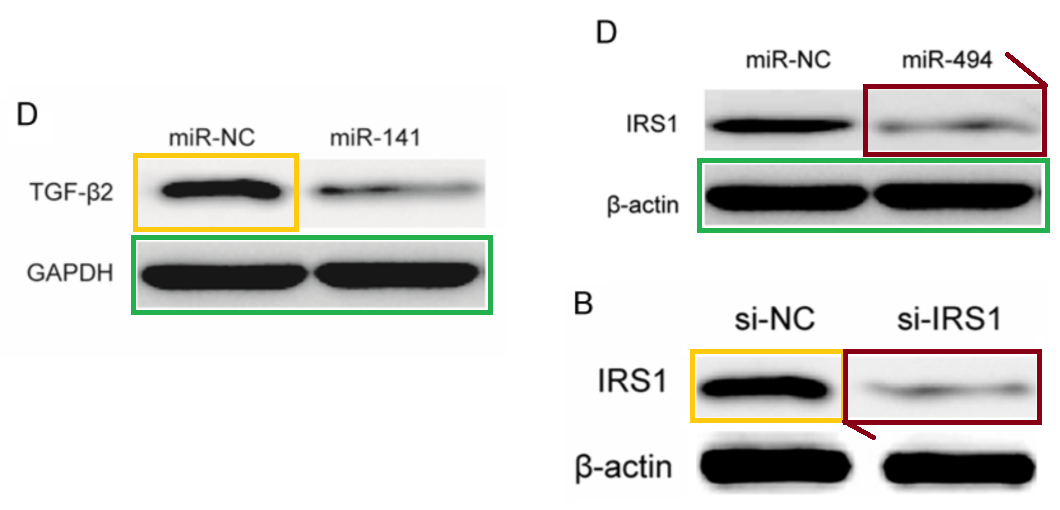
The bands of “Group 2” are sleek and elegant, like a Brancusi bird, and cry out to be cast in bronze. Manifestations include [left] Fig 3C from Ge et al (2016) (“Western blot analysis of uPA, MMP-2 and MMP-9 protein expression following transfection with miR-497 mimic or miR-control. GAPDH was used as an internal control“); and [right] Figs 3C,E from Liu et al (2015) [15] (“The … (C) protein expression levels of PTEN were detected in the HCC cells after transfection with miR-494 mimic or inhibitor. The PTEN protein expression level was detected in… (E) HCC cell lines by western blotting“).

I won’t prepare a comprehensive montage of these appearances for they feature in 30 of the papers in the corpus. Two bands are preferred loading controls, with substantive bands formed by cutting and splicing protein blobs that include the ‘decoy duck’ and the ‘swoosh’ and the ‘hockey-stick’.
Group-3 has only been sighted 22 times. It includes four different loading controls, but these often co-occur in the same papers, and they share the elements of their accompanying substantive bands, so there is no need to separate them in this taxonomy that no-one asked for.
Western-Blot groups 4, 5/6 and 7 have only shown up 14, 11 and 14 times respectively, and I’m getting bored, so we can skip them and move along. Group-4 are the ‘skate suite’ to me, on account of the appearance of one of the blobs. Group-5/6 are an ugly mishmash, featuring in layer-cake Figures like high-rise apartment blocks. Finally, Group-7 is all about furtive snakes. There are other repeated WBs that manifest twice only… but you get the picture, so let’s not dwell on them, and move on instead to some solar photography during peak sun-spot season. Or, as the case may be, colony-formation assay plates.
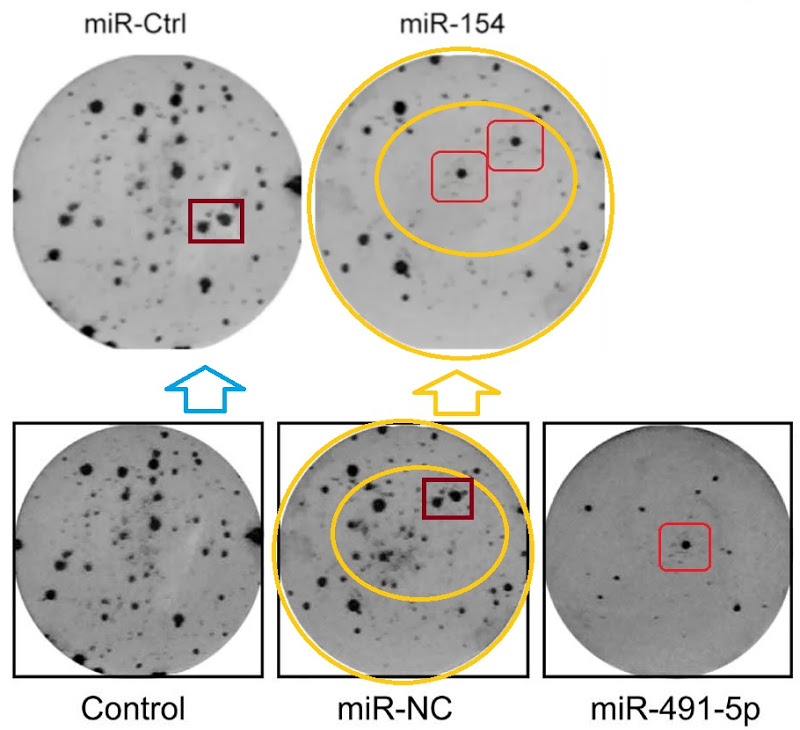
The Department of Unexpected Similarities is proud to bring you Fig 2 from Zhao et al (2015) [14] (“Effects of miR-491-5p overexpression on the colony formation of cervical cancer cells were determined“)…
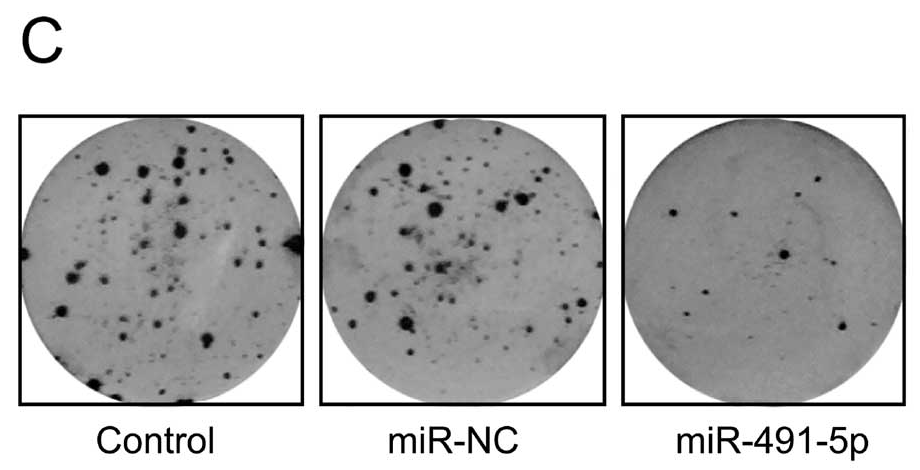
… also Fig 3E from Song et al (2015) [16] (“Determination of the capacity for colony formation of HeLa cells using a colony formation assay after transfection with miR-133a or miR-Ctrl“); and Fig 2C from [24] (“The capacity of colony formation was determined by colony formation assay in SKOV3 cells after transfection with miR-494 or miR-NC)“.
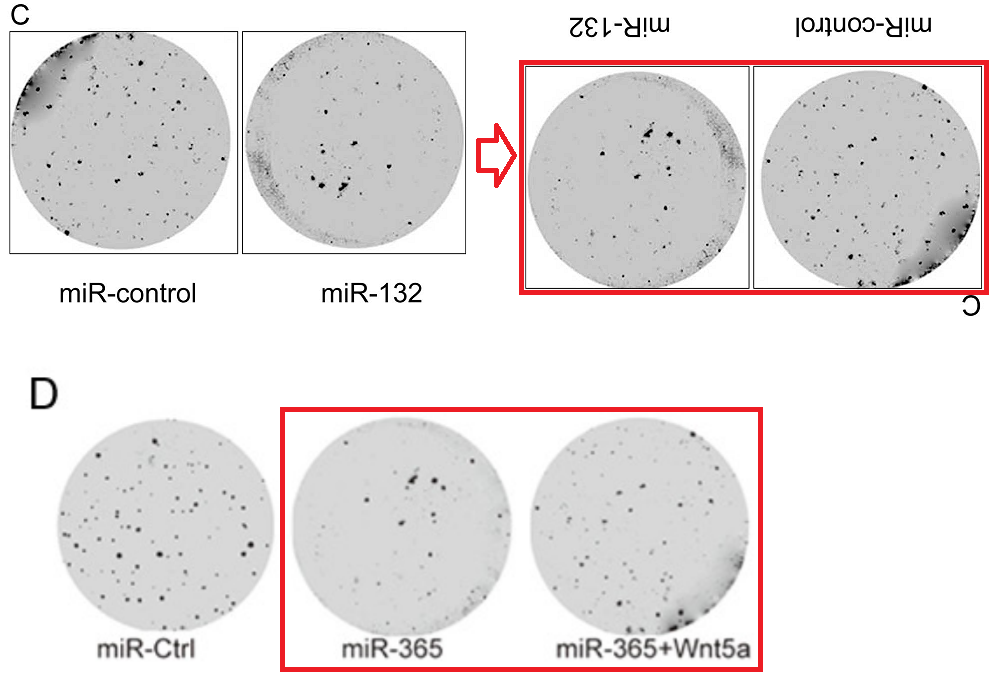
Then the studio became more ambitious, resulting in Fig 2C from Xu et al (2016) [27] (“Cell colony formation was determined in MCF-7 cells transfected with miR-154 mimic or miR-Ctrl”).
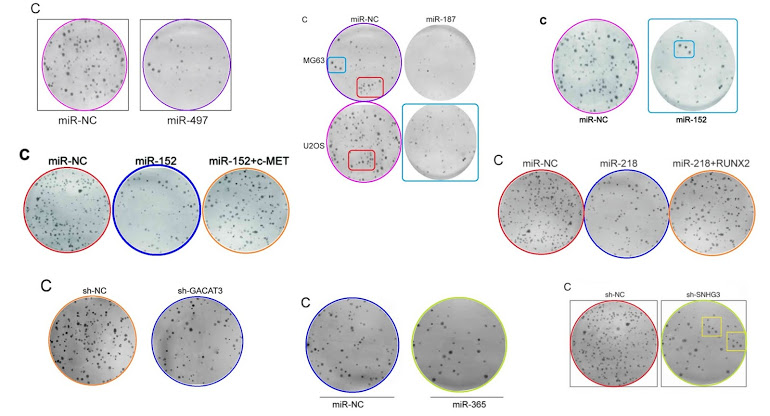
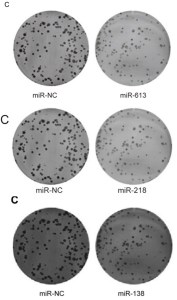 Also, for the sake of completeness: Figs 2C from Li et al (2016) [31]; 2C from [40]; and 2C from Pang et al (2017) [35]. But retouching colony-formation plates is addictive and you require stronger and stronger doses. Left-to-right, and top-to-bottom, Figs 2C from [29], 2C from Fei et al (2016) [30], and 2C from Li et al (2017a) [41; Figs 5C from [41] and 5C from [40]; Figs 2C from Feng et al (2018) [38], 2C from Liu et al (2017) [33], and 2C from [51].
Also, for the sake of completeness: Figs 2C from Li et al (2016) [31]; 2C from [40]; and 2C from Pang et al (2017) [35]. But retouching colony-formation plates is addictive and you require stronger and stronger doses. Left-to-right, and top-to-bottom, Figs 2C from [29], 2C from Fei et al (2016) [30], and 2C from Li et al (2017a) [41; Figs 5C from [41] and 5C from [40]; Figs 2C from Feng et al (2018) [38], 2C from Liu et al (2017) [33], and 2C from [51].
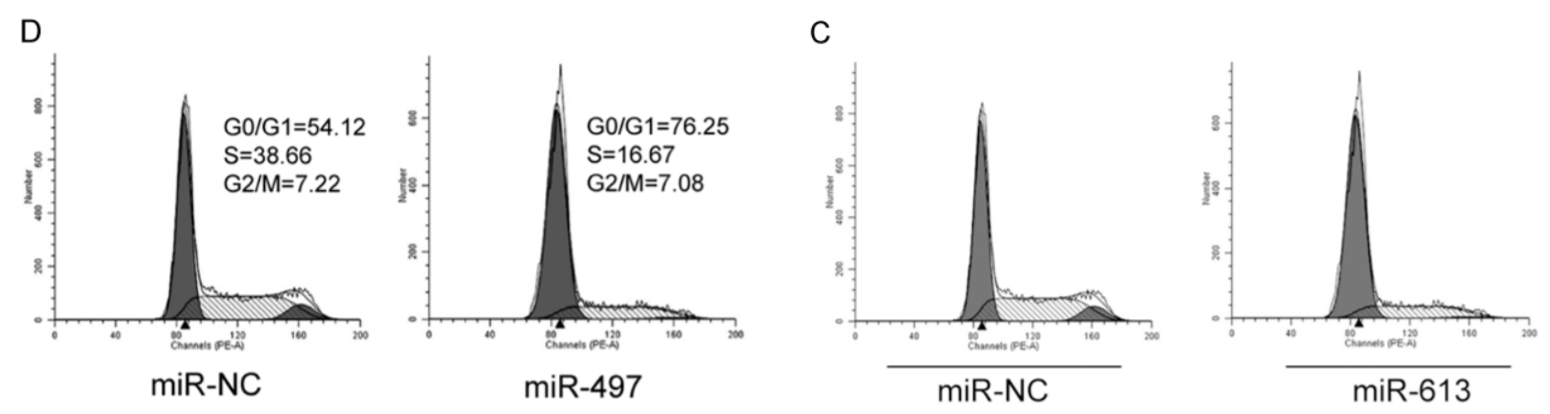
Another form of anapests to include in Figures as a visual claim of authenticity are the Flow-Cytometry histograms: “See, we quantified some treatment’s effect of disrupting the cell-division cycle in carcinoma cell-lines!” They are reproduced at roughly postage-stamp scale, and one can understand why the studio wasted no time customising new ones for each paper (although these are the easiest kind of “beautiful evidence” to fake), for their role is subliminal, with no expectation that readers will look at them. Instead we see double use.
Or triple use, and you can be forgiven for misreading “G0/G1” as “GIGO“.
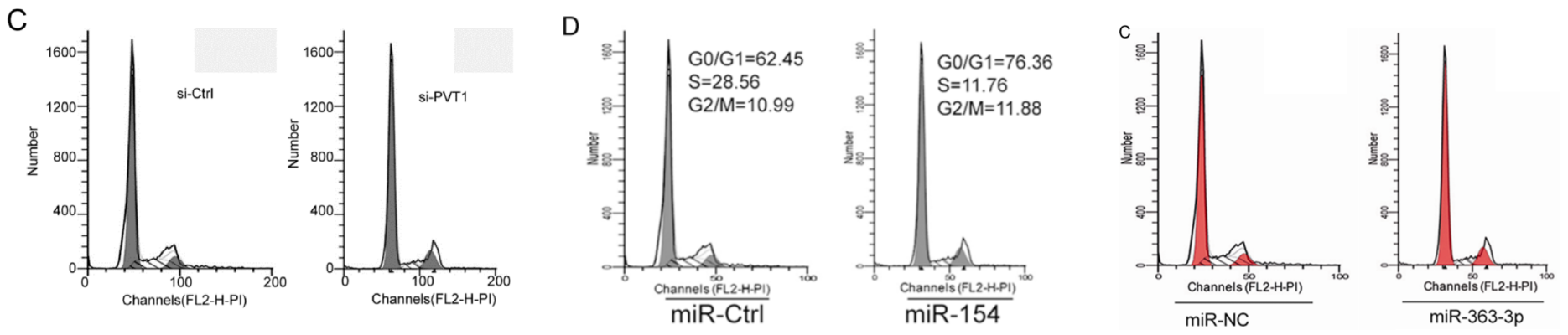
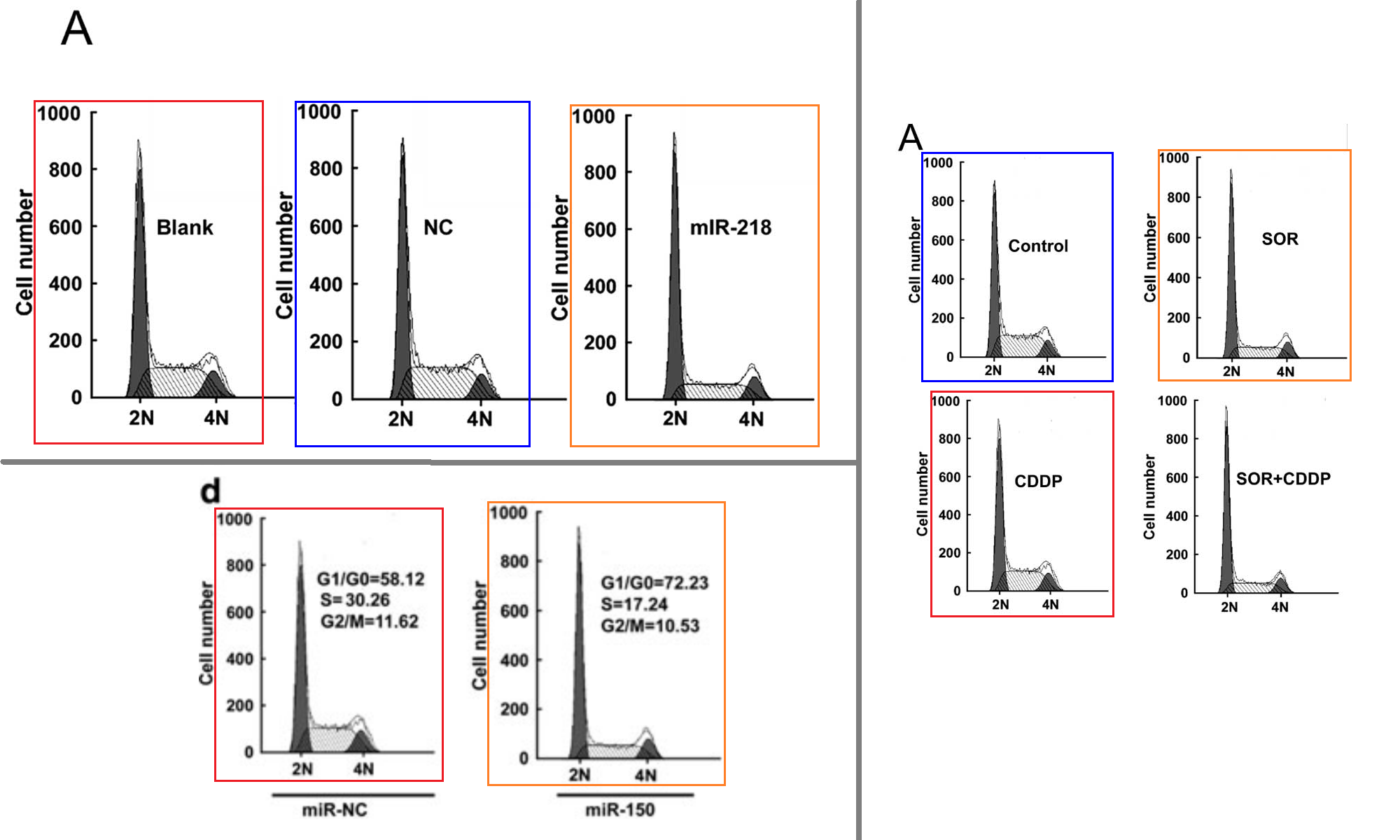
Or the same histograms straddling 12 papers.
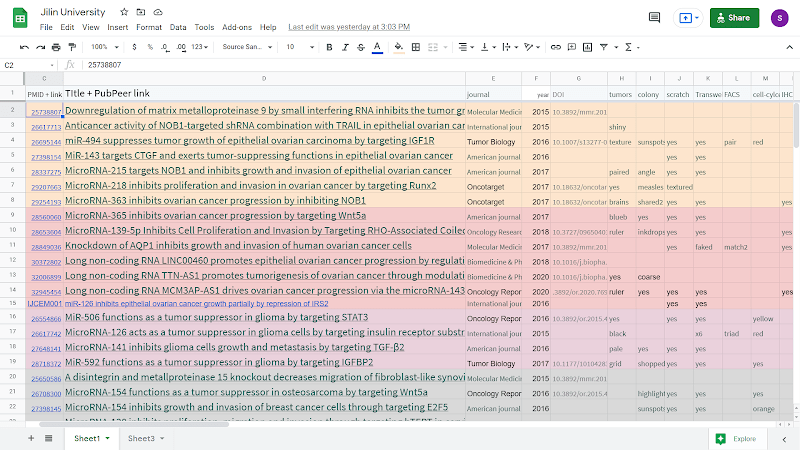
I want to stress that these suites of western blots, colony-plates and tumors are not specific to single departments or research teams. When I organised the spreadsheet entries into co-author networks, one network from the Thyroid Surgery department turned out to draw upon the entire gamut of anapests to improve treatment for papillary thyroid cancer. Other clusters from the Neurosurgery Department and the Gynecology Department sampled the entire gamut to improve treatment for glioma and ovarian cancer respectively.
The typical departmental network has one or two core members: the Head of Department or a prominent professor, I surmise – with the rest of the credit for publication spread around a shifting cast. The coveted title of “PubPeer Star” is shared between Zhihong Li and Wei Zhang, with 15 PubPeerified contributions to fantasy-fiction Thoracic Surgery and Hepatopancreatobiliary Surgery respectively. To be scrupulously fair, most of their CVs are clinical reports, not relevant here. My exercise in Network Analysis was slightly complicated by the discovery that some authors appear to be technical support: they are not confined to single research programs by the branches of the departmental hierarchy, and join up with otherwise-separate teams.
A second cluster of thyroid-cancer fairy-tales woven out of these visual elements proved to be the work of a network in the Department of Nuclear Medicine. One Qingjie Ma co-authored six of these fabulations, and was Corresponding Author for four of them. We read elsewhere that
“Professor Ma Qingjie is the director, chief physician, professor, and doctoral supervisor of the Department of Nuclear Medicine, China-Japan Friendship Hospital of Jilin University. Engaged in clinical work for more than 30 years. He has expertise and innovation in the personalized treatment of human thyroid diseases, various types of pediatric hemangioma, and certain tumor diseases; for multimodal molecules such as PET/CT and SPECT/CT He has deep attainments in imaging research and tumor-targeted drug therapy. He was named “Bethune Famous Doctor”, “Bethune Famous Teacher”, “Bethune Medical Worker”, “Expert with Outstanding Contributions” in Changchun City, Jilin University. In 2019, he won the Chinese Nuclear Medicine Physician Award.“
To sum up, this whole body of papers offers nothing but fabulation and flim-flam, and it does the research community no credit that they’ve attracted an average of 28 citations each (according to the duplicate-prone, inflated estimates of Goofle Scholar). The citation count peaks at 115 for [15], so I have read and re-read that work on the theory that there must be some underlying truth, but it stubbornly persists in remaining total wibble. Some of that number will be reciprocated hat-tips and professional courtesy from other fakers in this secondary literature, but not all of it.
Papers in Spandidos journals from the Jilin Studio tapered off in 2018. I initially wondered whether the studio is inactive now, but no… this only reflects a shift in preferred target journal, to Biomedicine & Pharmacotherapy from Elsevier now that it is part of the demi-monde literature. Perhaps the keepers of the Spandidos stable-gates were responding to submissions with an increasingly skeptical reception (suspicion is always corrosive in a relationship). Perhaps the administration at Jilin University were keen to have more people reading and believing the faculty’s Stakhanovite production of bullshit, and Elsevier offered a higher impact factor. One can only speculate.
To be scrupulously fair, other torrents of fake papers pouring forth from Jilin University were provided by undeniable papermills and do not have this studio’s hallmarks. A sixth of the prodigious output of the ‘Tadpole Papermill‘ was commissioned by staff at Jilin’s ancillary hospitals. Jilin shows up as a customer of other papermills as well. There may even be some genuine research happening there. I suspect, though, that anyone trying to conduct actual studies would be disadvantaged in the greasy-pole-climb of academic advancement by the time requirements of ‘experiments’ and the expenses of ‘laboratory’ and ‘equipment’.
Meanwhile the first retractions of the CSU fake papers have already begun, spicing up the interminable lists of newer fake papers. Each retraction note credits “a concerned reader” for drawing the bullshit “to the Editors’ attention”, whereupon the Editors were shocked, SHOCKED to find gambling going on. Which is to say, the Spandidos management read PubPeer but don’t want to acknowledge its existence. A great culling of Jilin crap cannot be far away.

Publishers in the demi-monde are willing to retract papers, as a pawn sacrifice or an ablative heat-shield, once someone else’s peer-reviewing shows them up as plagiarised or fabricated. Perhaps ‘delighted’ is more apt than ‘happy’ as each retraction shows that the conduit is just like a real journal in terms of integrity standards, while diverting attention away from the question, “why did you accept such abject tosh in the first place?” Also the publisher keeps the signatories’ money, so retraction costs them nothing.
SOURCES
- “miR 222 regulates sorafenib resistance and enhance tumorigenicity in hepatocellular carcinoma“, KAI LIU , SONGYANG LIU , WEI ZHANG , BAI JI , YINGCHAO WANG , YAHUI LIU (2014). International Journal of Oncology.
- “Upregulation of GRIM-19 suppresses the growth of oral squamous cell carcinoma in vitro and in vivo“, MINGHE LI , ZHIHONG LI , CHONGYANG LIANG , CHENGMIN HAN , WEI HUANG , FEI SUN (2014). Oncology Reports.
- “Knockout of ADAM10 enhances sorafenib antitumor activity of hepatocellular carcinoma in vitro and in vivo“, Wei Zhang , Songyang Liu , Kai Liu , Bai Ji , Yingchao Wang , Yahui Liu (2014). Oncology Reports.
- “Synergistic effects of co-expression plasmid based ADAM10-specific siRNA and GRIM-19 on hepatocellular carcinoma in vitro and in vivo“, Songyang Liu , Wei Zhang , Kai Liu , Yingchao Wang , Bai Ji , Yahui Liu (2014). Oncology Reports.
- “Synthetic miR-145 mimic inhibits multiple myeloma cell growth in vitro and in vivo“, Qi Zhang , Weiqun Yan , Yang Bai , Hao Xu , Changhao Fu , Wenwen Zheng , Yingqiao Zhu , Jie Ma (2014). Oncology Reports.
- “miR-218 suppresses tumor growth and enhances the chemosensitivity of esophageal squamous cell carcinoma to cisplatin“, Hang Tian , Lei Hou , Yu-Mei Xiong , Jun-Xiang Huang , Ying-Jun She , Xiao-Bao Bi , Xing-Rong Song (2015). Oncology Reports.
- “Combined treatment of XIAP-targeting shRNA and celecoxib synergistically inhibits the tumor growth of non small cell lung cancer cells in vitro and in vivo“. HONG ZHANG , ZHIHONG LI , KAIZHONG WANG , PING REN (2015). Oncology Reports.
- “Silencing XIAP suppresses osteosarcoma cell growth, and enhances the sensitivity of osteosarcoma cells to doxorubicin and cisplatin“, YANG QU , PENG XIA , SHANYONG ZHANG , SU PAN , JIANWU ZHAO (2015). Oncology Reports.
- “Silencing NOB1 enhances doxorubicin antitumor activity of the papillary thyroid carcinoma in vitro and in vivo“, JIA LIU , BING-FEI DONG , PEI-SONG WANG , PEI-YOU REN , SHUAI XUE , Xiao-Νan ZHANG , ZHE HAN , GUANG CHEN (2015). Oncology Reports.
- “A disintegrin and metallproteinase 15 knockout decreases migration of fibroblast-like synoviocytes and inflammation in rheumatoid arthritis“, JINLIANG GAO , WEI ZHENG , LIMING WANG , BAILIN SONG (2015). Molecular Medicine Reports.
- “Synergistic growth inhibition by sorafenib and cisplatin in human osteosarcoma cells“, QU YANG , SHANYONG ZHANG , MINGYANG KANG , RONGPENG DONG , JIANWU ZHAO (2015). Oncology Reports.
- “Downregulation of matrix metalloproteinase 9 by small interfering RNA inhibits the tumor growth of ovarian epithelial carcinoma in vitro and in vivo“, FENGJUN GUO , JINGYAN TIAN , MANHUA CUI , MEIRU FANG , LIN YANG (2015). Molecular Medicine Reports.
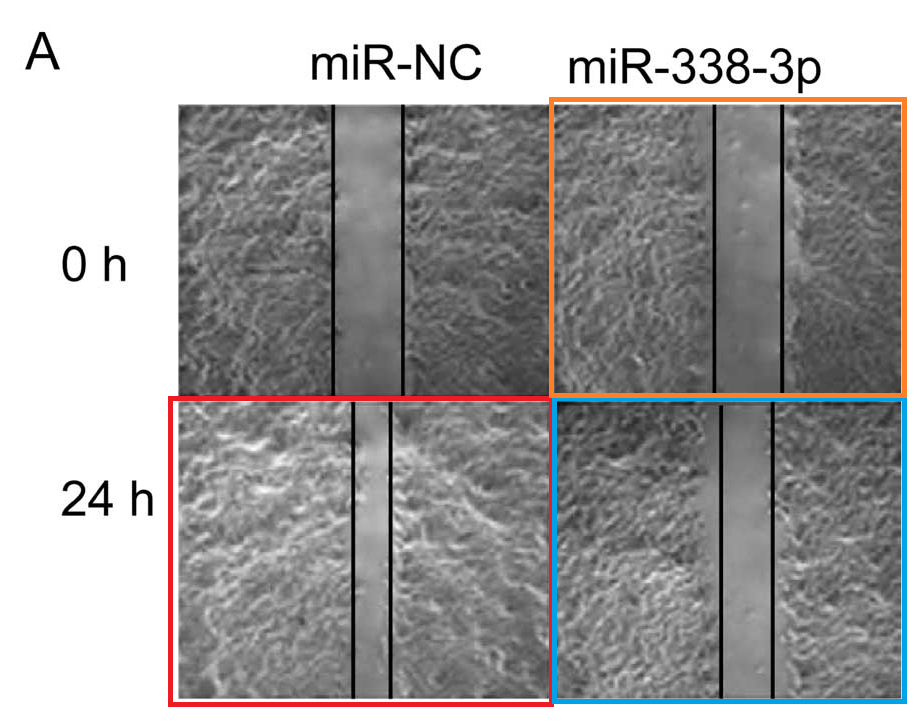
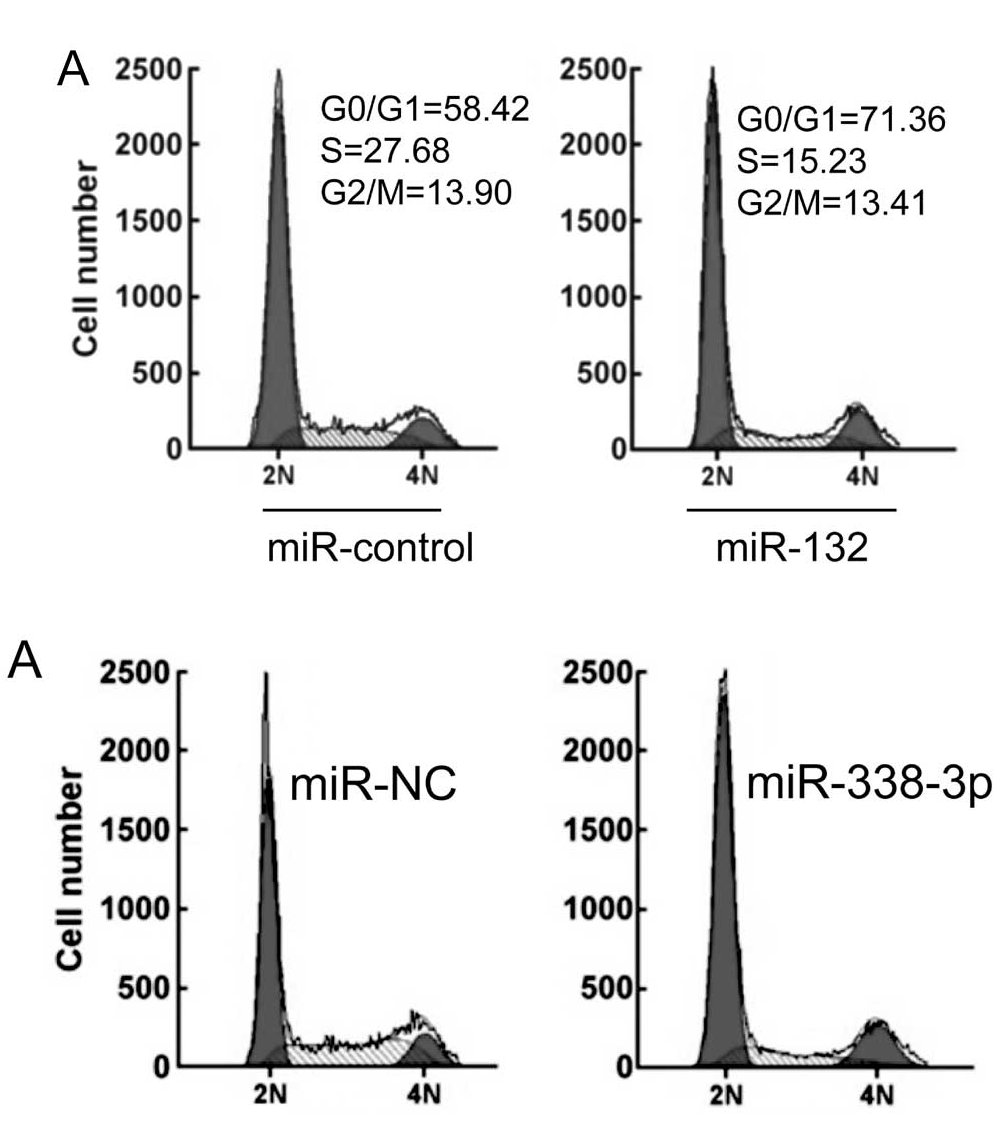
- “MicroRNA-338-3p suppresses tumor growth of esophageal squamous cell carcinoma in vitro and in vivo“, XINYU LI , ZHIHONG LI , GUIYUN YANG , ZHENXIANG PAN (2015). Molecular Medicine Reports.
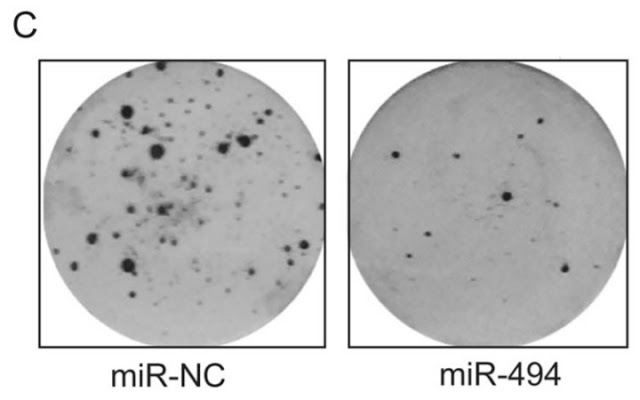
- “MicroRNA-491-5p suppresses cervical cancer cell growth by targeting hTERT“, Qiang Zhao, Ying-Xian Zhai, Huan-Qiu Liu, Ying-Ai Shi, Xin-Bai Li (2015). Oncology Reports.
- “miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN“, Kai Liu, Songyang Liu, Wei Zhang, Baoxing Jia, Ludong Tan, Zhe Jin, Yahui Liu (2015). Oncology Reports.
- “miR-133a inhibits cervical cancer growth by targeting EGFR“, Xuesong Song, Bo Shi, Kexin Huang, Wenjie Zhang (2015). Oncology Reports.
- “Knockdown of ADAM10 inhibits migration and invasion of fibroblast-like synoviocytes in rheumatoid arthritis“, DAN LI, ZHITAO XIAO, GANG WANG, XIANJI SONG (2015). Molecular Medicine Reports.
- “MiR-132 inhibits cell proliferation, invasion and migration of hepatocellular carcinoma by targeting PIK3R3“, KAI LIU, XINGLIANG LI, YUCHEN CAO, YUANYUAN GE, JIANMENG WANG, BO SHI (2015). International Journal of Oncology.
- “MicroRNA-338-3p functions as tumor suppressor in breast cancer by targeting SOX4“, YING JIN, MIN ZHAO, QIAN XIE, HONGYAN ZHANG, QING WANG, QINGJIE MA (2015). International Journal of Oncology.
- “MicroRNA-128 suppresses cell growth and metastasis in colorectal carcinoma by targeting IRS1“, Lan Wu, Bo Shi, Kexin Huang, Guoyu Fan (2015). Oncology Reports.
- “miR-29a suppresses growth and metastasis in papillary thyroid carcinoma by targeting AKT3“, Rui Li, Jia Liu, Qun Li, Guang Chen, Xiaofang Yu (2016). Tumor Biology.
- “miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor“, Xiaoli Pang, Kexin Huang, Qianqian Zhang, Yujiao Zhang, Junqi Niu (2015). Oncology Reports.
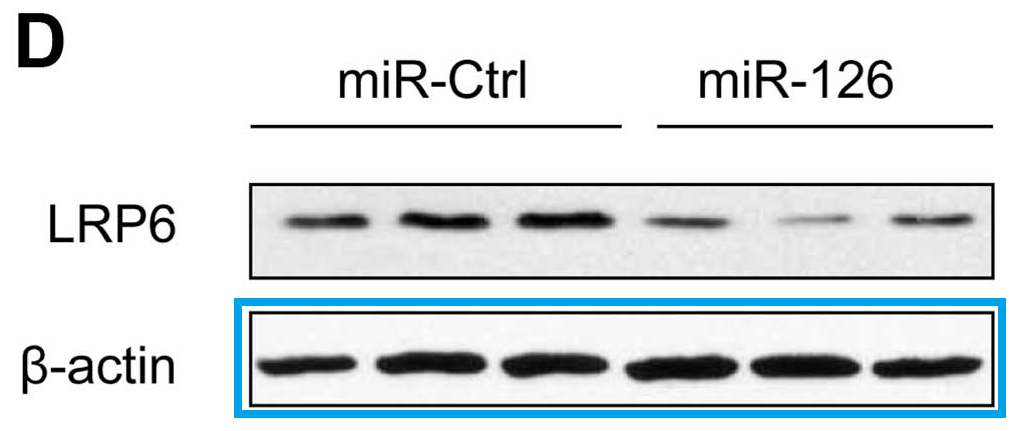
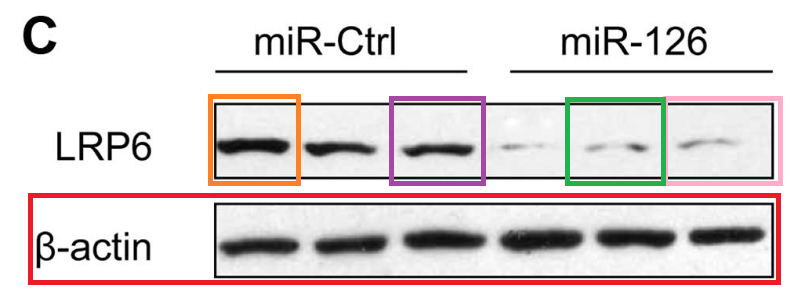
- “MicroRNA-126 acts as a tumor suppressor in glioma cells by targeting insulin receptor substrate 1 (IRS-1)“, Yongxin Luan, Ling Zuo, Shuyan Zhang, Guangming Wang, Tao Peng (2015). International Journal of Oncology.
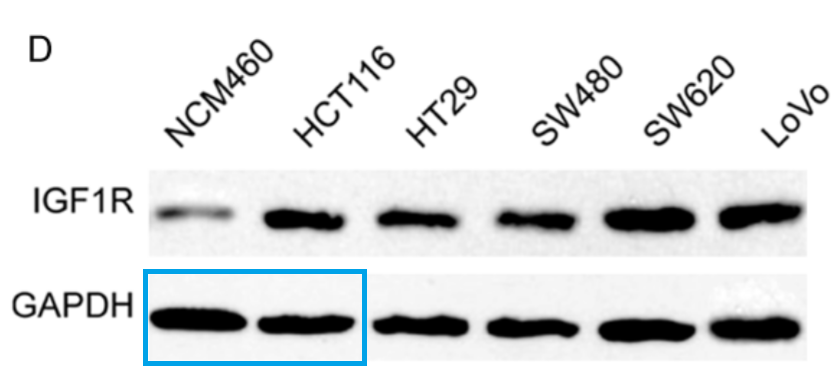
- “miR-494 suppresses tumor growth of epithelial ovarian carcinoma by targeting IGF1R“, Na Li, Xiaosu Zhao, Lufei Wang, Shi Zhang, Manhua Cui, Jin He (2016). Tumor Biology.
- “Combined RNAi targeting human Stat3 and ADAM9 as gene therapy for non-small cell lung cancer“, LIANG CHANG, FANGCHAO GONG, HONGFEI CAI, ZHIHONG LI, YOUBIN CUI (2016). Oncology Letters.
- “miR-449a inhibits proliferation and invasion by regulating ADAM10 in hepatocellular carcinoma“, Songyang Liu, Kai Liu, Wei Zhang, Yingchao Wang, Zhe Jin, Baoxing Jia, Yahui Liu (2016). American journal of translational research.
- “MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5“, Hui Xu, Dan Fei, Shan Zong, Zhimin Fan (2016). American journal of translational research.
- “MicroRNA-107 inhibits tumor growth and metastasis by targeting the BDNF-mediated PI3K/AKT pathway in human non-small lung cancer“, Huan Xia, Yang Li, Xiaohong Lv (2016). International Journal of Oncology.
- “MicroRNA-497 inhibits thyroid cancer tumor growth and invasion by suppressing BDNF“, Peisong Wang, Xianying Meng, Yan Huang, Zhi Lv, Jia Liu, Guimin Wang, Wei Meng, Shuai Xue, Qiang Zhang, Pengju Zhang, Guang Chen (2017). Oncotarget.
- “MicroRNA-187 exerts tumor-suppressing functions in osteosarcoma by targeting ZEB2“, Dan Fei, Kunchi Zhao, Hongping Yuan, Jie Xing, Dongxu Zhao (2016). American journal of cancer research.
- “MicroRNA-613 suppresses proliferation, migration and invasion of osteosarcoma by targeting c-MET“, Xinyu Li, Xufang Sun, Jing Wu, Zhihong Li (2016). American journal of cancer research.
- “MicroRNA-497 suppresses cell proliferation and induces apoptosis through targeting PBX3 in human multiple myeloma“, Tianhua Yu, Xuanhe Zhang, Lirong Zhang, Yali Wang, Hongjuan Pan, Zhihua Xu, Xiaochuan Pang (2016). American journal of cancer research.
- “miR-365 targets ADAM10 and suppresses the cell growth and metastasis of hepatocellular carcinoma“, Yahui Liu, Wei Zhang, Songyang Liu, Kai Liu, Bai Ji, Yingchao Wang (2017). Oncology Reports.
- “MicroRNA-365 inhibits ovarian cancer progression by targeting Wnt5a“, Yanli Wang, Chunling Xu, Yun Wang, Xiaomeng Zhang (2017). American journal of cancer research.
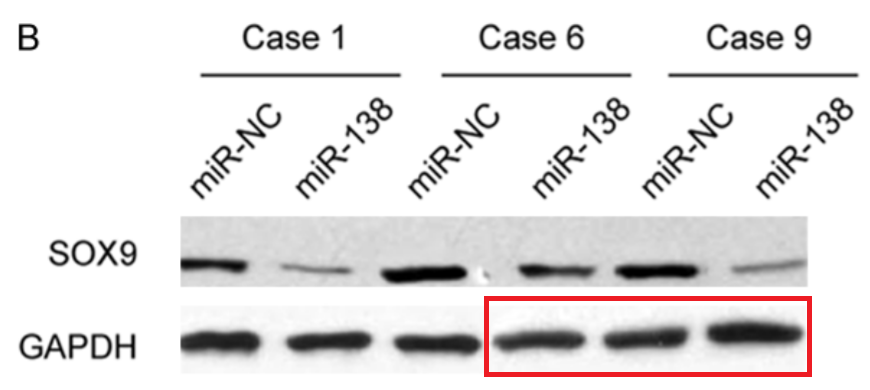
- “miR-138 inhibits gastric cancer growth by suppressing SOX4“, Lei Pang, Bai Li, Baisong Zheng, Liang Niu, Liang Ge (2017). Tumor Biology.
- “MicroRNA-25 suppresses proliferation, migration, and invasion of osteosarcoma by targeting SOX4“, Bingpeng Chen, Jingjing Liu, Ji Qu, Yang Song, Yuxiang Li, Su Pan (2017). Tumor Biology.
- “miR-217 inhibits proliferation, migration, and invasion via targeting AKT3 in thyroid cancer“, Yuanqiang Lin, Kailiang Cheng, Tongtong Wang, Qian Xie, Minglong Chen, Qianqian Chen, Qiang Wen (2017). Biomedicine & Pharmacotherapy.
- “LncRNA GACAT3 promotes gastric cancer progression by negatively regulating miR-497 expression“, Li Feng, Yonggang Zhu, Yunxin Zhang, Min Rao (2017). Biomedicine & Pharmacotherapy.
- “Function of miR-212 as a tumor suppressor in thyroid cancer by targeting SIRT1“, Dandan Li, Lin Bai, Tongtong Wang, Qian Xie, Minglong Chen, Yantao Fu, Qiang Wen (2017). Oncology Reports.
- “MicroRNA-218 inhibits proliferation and invasion in ovarian cancer by targeting Runx2“, Na Li, Lufei Wang, Guangyun Tan, Zhiheng Guo, Lei Liu, Ming Yang, Jin He (2017). Oncotarget.
- “Function of miR 152 as tumor suppressor in oral squamous cell carcinoma cells by targeting c MET“, Minghe Li, Zhihong Li, Xue Wang, Yumei Wang, Cong Zhao, Lei Wang (2017). Oncology Reports.
- “miR 152 inhibits rheumatoid arthritis synovial fibroblast proliferation and induces apoptosis by targeting ADAM10“, Jialong Guo, Juan Du, Dan Fei, Jie Xing, Jinxiang Liu, Honghua Lu (2017). International journal of molecular medicine.
- “MicroRNA-361-5p inhibits papillary thyroid carcinoma progression by targeting ROCK1“, Rui Li, Bingfei Dong, Zhengmin Wang, Tao Jiang, Guang Chen (2018). Biomedicine & Pharmacotherapy.
- “Knockdown of CREB1 promotes apoptosis and decreases estradiol synthesis in mouse granulosa cells“, Pengju Zhang, Jun Wang, Hongyan Lang, Weixia Wang, Xiaohui Liu, Haiyan Liu, Chengcheng Tan, Xintao Li, Yumin Zhao, Xinghong Wu (2018). Biomedicine & Pharmacotherapy.
- “Long Noncoding RNA FER1L4 Suppresses Tumorigenesis by Regulating the Expression of PTEN Targeting miR-18a-5p in Osteosarcoma“, Dan Fei, Xiaona Zhang, Jinxiang Liu, Long Tan, Jie Xing, Dongxu Zhao, Yang Zhang (2018). Cellular Physiology & Biochemistry.
- “LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p“, Songyang Liu, Wei Zhang, Kai Liu, Yahui Liu (2018). Biomedicine & Pharmacotherapy.
- “Long noncoding RNA HOXA-AS2 promotes non-small cell lung cancer progression by regulating miR-520a-3p“, Yunpeng Liu, Xingyu Lin, Shiyao Zhou, Peng Zhang, Guoguang Shao, Zhiguang Yang (2019). Bioscience Reports.
- “Long non‐coding RNA deleted in lymphocytic leukaemia 1 promotes hepatocellular carcinoma progression by sponging miR‐133a to regulate IGF‐1R expression“, Wei Zhang, Songyang Liu, Kai Liu, Yahui Liu (2019). Journal of Cellular and Molecular Medicine.
- “Long noncoding RNA AFAP1-AS1 promotes osteosarcoma progression by regulating miR-497/IGF1R axis“, Dan Fei, Xiaona Zhang, Yang Lu, Long Tan, Mingzhu Xu, Yang Zhang (2019). American journal of translational research.
- “Long non coding RNA MCM3AP AS1 drives ovarian cancer progression via the microRNA 143 3p/TAK1 axis“, Jihong Wen, Shumei Han, Man Cui, Yanli Wang (2020). Oncology Reports.
- “lncRNA SNHG3 promotes breast cancerprogression by acting as a miR 326 sponge“, Haipeng Zhang, Na Wei, Wei Zhang, Lishennan Shen, Rongbo Ding, Qian Li, Simin Li, Ye Du (2020). Oncology Reports.

Get For Better Science delivered to your inbox.
Make a one-time donation
Make a monthly donation
Choose an amount
Or enter a custom amount
Your generous patronage of my journalism will be most appreciated!
Your contribution is appreciated.
DonateDonate monthly















The retractions will continue until morale improves!
31 May. “Following the publication of this paper, it was drawn to the Editors’ attention by a concerned reader that certain of the western blotting data shown in Figs. 4C, 5B and 6D were strikingly similar to data appearing in different form in other articles by different authors”.
2 June. “Following the publication of this paper, it was drawn to the Editors’ attention by a concerned reader that the western blotting data in Fig. 5c were strikingly similar to data appearing in different form in other articles by different authors at different research institutes.”
For some reason they cling to the pretense that it’s always “a concerned reader” who called the frauds to the Editors’ attention, when they could simply admit that they read PubPeer.
the provenance of any material in this whole genre that is not simply repeated with rearrangement and manipulation
Here’s a retraction from Dove Press, 3 June.”The Editor and Publisher of OncoTargets and Therapy wish to retract the published article. The journal was notified of alleged image manipulation relating to Figure 3B”…
In this case, the papermillers had made the mistake of grabbing some random Transwell image from an earlier paper to illustrate their fantasy, without realising that the earlier paper was itself a papermill product, so the same image appears repeatedly in other papers from the same source. Not a good look.
LikeLike
Also Zhen Tian got promoted despite a retraction and other paper mill fraud he published
Ah yes, “microRNA-493 inhibits tongue squamous cell carcinoma oncogenicity via directly targeting HMGA2”. That was from another papermill… or perhaps a known papermill, using a new template to churn out non-coding-RNA papers with the words “directly targeting” in the title (or just “targeting”).
https://pubpeer.com/search?q=directly+targeting
LikeLike
Many of the articles found in https://pubpeer.com/search?q=microRNA+%22directly+targeting%22 use the same images. Its so lazy.
LikeLike
Yes indeed, they use flow-cytometry plots drawn from a rather small collection.
https://docs.google.com/spreadsheets/d/1X401JM6CcpTSkbKsPsOl20pRKQ-ns1S_J5pTwtBjqPw/edit#gid=0
LikeLike
Perhaps the paper mills are a PRC disinformation campaign to slow down the advances of non-Chinese researchers.
LikeLike
China emulates and does better. They figured out what the real purpose of the western science publishing output was, and now took over. It’s not like Elsevier is complaining.
LikeLike
“Think of all the many honest Chinese scientists in Jilin and elsewhere who will not get any promotions or grants, or even lose their jobs, all because they refused to commit fraud.”
Name two – looks to me like the whole country is at it.
LikeLike
I can’t name any because they were sacked for not publishing enough.
LikeLike
I heard that one of Jilin University hospitals recently had a meeting talking about their frauds had been investigated and disclosed by outsiders, guess they meant us. The leadership was saying that, as a hospital academically affiliated to a good university, their doctors are obligated to publish, that’s for sure, otherwise their doctors are no different from those from other non-academic hospitals. But they are aware of the time/effort restriction for those doctors who have heavy clinical load, and they are talking about probably (or hopefully) to release doctors from some clinical hours to do their research, rather than having them had to purchase from others.
LikeLike
Maybe some ethics training might be in order? Here’s another thought: they seem to have more ready money than time, since they can buy papers but not do actual research. Why not fine everyone caught buying papers a sufficient amount that would act as a disincentive?
LikeLike
Pingback: The Master of the String-of-Sausages – For Better Science
Pingback: The papermills of my mind – For Better Science