The brave women and men of American Society of Plant Biologists (ASPB) did a bold step towards ensuring research integrity in plant sciences. Fraud is no more, honesty restored, science and not Photoshop shall serve the greater good of agriculture and feed the humanity from now on. All this will become reality because Leonid Schneider has been blocked on Twitter by ASPB and their community site Plantae. Well, transiently, but still. It might have been a warning to me!
It’s not like I didn’t deserve getting blocked. Disinformation flow had to be stopped, about some shenanigans going on at ASPB’s flagship journal The Plant Cell, responsible for which were their own editors, including the newly appointed Editor-in-Chief Sally Assmann. These are the members and top-authors of ASPB whom I must thank for my tough but fair Twitter lesson:

One of these brave ASPB heroes whose honour needed defending is the gentleman I marked with a frame: Gary Stacey , Endowed Professor of Soybean Biotechnology at University of Missouri in USA. He is also ASPB member and one of recognised “ASPB top authors”. You can imagine the industrial scale cash which the soy and legume expert Stacey receives from agricultural multinationals, in recognition for his outstanding research which is now being questioned. And I trolled him on Twitter.
The Stacey affair started rather small, with relatively minor issues in his papers. It soon escalated as readers began to send me evidence of data manipulations for which no innocent explanations were possible, some of it was posted also on PubPeer. Yet it seems, Stacey’s employer the University of Missouri closed the case already. My attempt to submit additional evidence was not answered. Stacey declared on Twitter that the university attested him a “clean bill of health”, the university did not deny that.
For example, look what Stacey lab in Missouri (which the boss runs together with his wife and former PhD student, Minviluz Stacey) published:

This Photoshop hack job saw the light of the day almost 20 years ago, as Figure 10 in this paper:

R. Bradley Day, Crystal B. McAlvin, John T. Loh, Roxanne L. Denny, Todd C. Wood, Nevin D. Young, Gary Stacey
Differential expression of two soybean apyrases, one of which is an early nodulin
Molecular plant-microbe interactions MPMI (2000) doi: 10.1094/mpmi.2000.13.10.1053
That Frankenstein Figure 10 apparently was already used as Figure 9D in same publication, and still it is not the only fabricated dataset there, look at Figure 7C:

Stacey’s reply to this evidence of blatant data fakery in his old paper was spot on. I need to get a life, apparently. Maybe. Or maybe Stacey’s own life ambition as plant researcher has always been to compete with Olivier Voinnet?
Technology has developed since, Adobe Photoshop kept issuing new editions and people became more skilled in the high art of illusions of scientific reality. The following gel collage looks more professional, it appeared in the ASPB journal Plant Physiology labelled in caps as “BREAKTHROUGH TECHNOLOGIES”, probably ironically.
Yaya Cui, Shyam Barampuram, Minviluz G. Stacey, C. Nathan Hancock, Seth Findley, Melanie Mathieu, Zhanyuan Zhang, Wayne A. Parrott, Gary Stacey
Tnt1 Retrotransposon Mutagenesis: A Tool for Soybean Functional Genomics
Plant Physiology (2013). DOI: 10.1104/pp.112.205369

In the next years, new Photoshop mastership levels were achieved in scientific publications. It seems that cheaters of today do not just manipulate figures for publication, they are even capable of fabricating pictures of original raw data in Photoshop, in case a journal editor or PubPeer commenter starts asking questions. Maybe this is why the journal eLife is still trying to figure out why the unrelated loading controls (shown below) look so strangely near-identical.
Yangrong Cao, Yan Liang, Kiwamu Tanak , Cuong T Nguyen, Robert P Jedrzejczak, Andrzej Joachimiak, Gary Stacey
The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1
eLife (2014) doi: 10.7554/elife.03766

That Cao et al eLife 2014 paper contains also a spliced-on gel lane in the Figure 5B (below, on the right), just as another paper from same time period (Garcia et al, Planta 2013) does, shown on the left. What was wrong with the original gel band so it needed replacing? Another stealthily spliced gel was spotted in Liao et al, New Phytologist 2017, but there Stacey explained on PubPeer that original data from less than 2 years ago proved unavailable, so he repeated the experiments to verify the result. Ho-hum.


The left-hand image from Garcia et al Planta 2013 is not on Pubpeer, only on Twitter, where Stacey stopped replying. But regarding the Cao et al eLife 2014 paper, he took a very bold approach of denial:
“We were made aware of the above comments on this paper via the eLife editors. In their email to us, +they already dismissed the comments on Fig. 6 & 7 as unfounded and, hence, we will not comment further. However, the eLife editors asked us to provide the original blot for Fig. 5.”
Stacey then provided some faint low-resolution “scan” purportedly showing the original gel for Figure 5. He then insinuated that the splicing he was accused of was actually an artefact introduced by the publisher when his original low-resolution figures were up-scaled to high-resolution when published online. Stacey’s conclusion:
“As one can see, there was no fudging of the figure as suggested.”
and
“The eLife editors have informed us that they are satisfied with our response so we now consider the issue closed.”
As I learned, eLife is still investigating, the issue is actually not closed. But the good news for Stacey might be that a potentially more problematic paper of his in Nature Communications is likely to be safe from even the most modest attempts at correction. That journal namely proved its new stance on research integrity by having refused doing anything at all about a manipulated paper by Karl Lenhard Rudolph, despite announcements from the German Leibniz Society and the DFG. Good for Rudolph, good for Stacey, heck, that sounds like the Nature Publishing Group is seriously competing with Cell Press.
This is Stacey’s paper, from 2017 and now it gets even weirder.
Dongqin Chen, Yangrong Cao, Hong Li, Daewon Kim, Nagib Ahsan, Jay Thelen, Gary Stacey
Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture
Nature Communications (2017) doi: 10.1038/s41467-017-02340-3
The PubPeer thread started with a luceferase luminescence figure which Stacey lab managed to detect with a fluorescence camera settings. It soon became much more exciting. Two issues were flagged: first, in Figure 2b a set of two HA immunoblot bands seems to rather neatly match its next neighbours on the same gel, while in the panel below, Myc-bands look suspiciously like two different exposures of same set of 3 bands, only horizontally flipped.
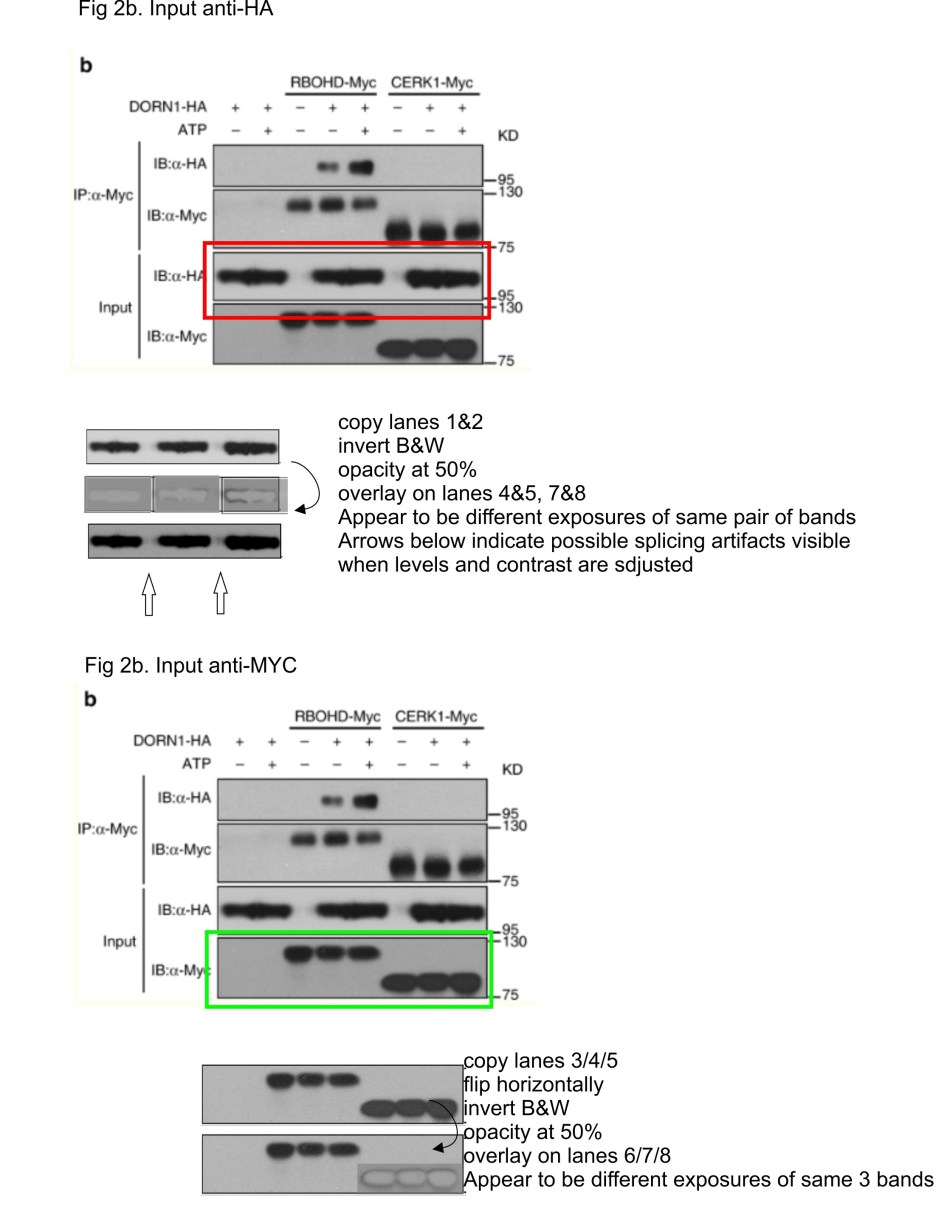
The other issue was that two different immunoblots, probed with same antibodies but with different samples loaded, not only suggested overlapping band shapes, even background splotches looked same. These splotches and dirt artefacts can happen during protein transfer from gel to western blot membrane, and if they happen, they are unique. All in all, it was rather obvious the images showed different exposures of the same blot membrane probed with GST antibody. A minor mistake of oversight?

Stacey’s replies on PubPeer however went into a different direction. Instead of taking an easy way out and admitting an error, he posted some raw data which he expected to prove all concerns wrong.

The raw data for Figure 2 reminds one of a different case, where another scientist, Timothy Chan of Memorial Sloan Kettering Cancer Center, uploaded “raw” data to PubPeer, yet the two bands still overlapped perfectly, something which cannot be explained by chance, only by application of Infinite Improbability Drive. It seems both Chan and Stacey labs are constantly struggling with western blots turning into sperm whales and bowls of petunias. There are of course alternative explanations, and it doesn’t let the principal investigator look good while waving such “raw data” pictures about.
But what Stacey uploaded for Figure 3a, still looks exactly like two exposures of the same western blot. Yet instead of finally admitting a small mistake, he even doubled down with same pictures in darker, still denying they show the same blot, and added as precaution:
“the net effect is that none of this has any real impact on the overall conclusions of the paper.”
Eventually, Stacey announced to be doing some “inquiries” into the mystery of how the gel bands happen to turn out looking identical in his lab. In this regard, there is also this other fresh paper in the ASPB journal Plant Physiology which Stacey coauthored with colleagues from Washington State University:
Diwaker Tripathi, Tong Zhang, Abraham J. Koo, Gary Stacey, Kiwamu Tanaka
Extracellular ATP Acts on Jasmonate Signaling to Reinforce Plant Defense
Plant Phys (2018) doi: 10.1104/pp.17.01477
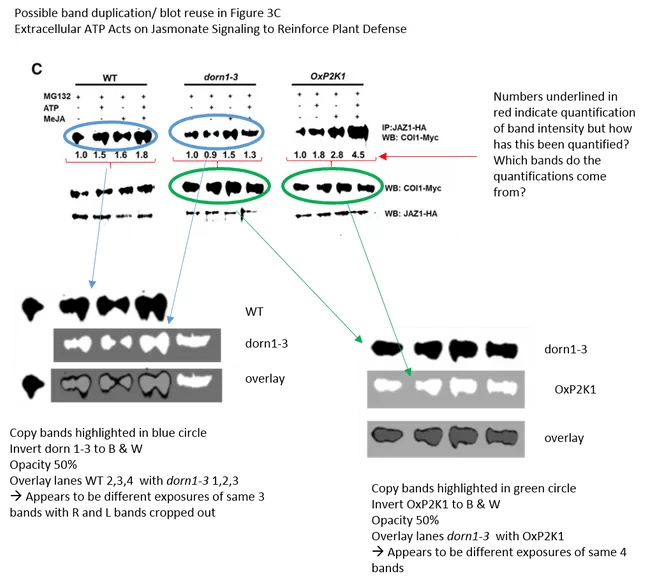
How did the gel band similarity there ever happen? Another magic coincidence? Or is this maybe the reason why the western blots in that paper lack all noise or background, to hide traces of data manipulation? Why does a journal accept figures of western blots looking like this, in 2018?
It is probably best to conclude this Stacey saga with a paper in Frontiers. It is also the freshest Stacey paper, merely a month old. If only Frontiers were able to muster enough decency and competence to ask for original data from their paying customers, and then act appropriately. Because in that paper, the soybean lab published some actual artwork.
Md Shakhawat Hossain, Nhung T. Hoang, Zhe Yan, Katalin Tóth, Blake C. Meyers, Gary Stacey
Characterization of the Spatial and Temporal Expression of Two Soybean miRNAs Identifies SCL6 as a Novel Regulator of Soybean Nodulation
Frontiers in Plant Science (2019) doi: 10.3389/fpls.2019.00475
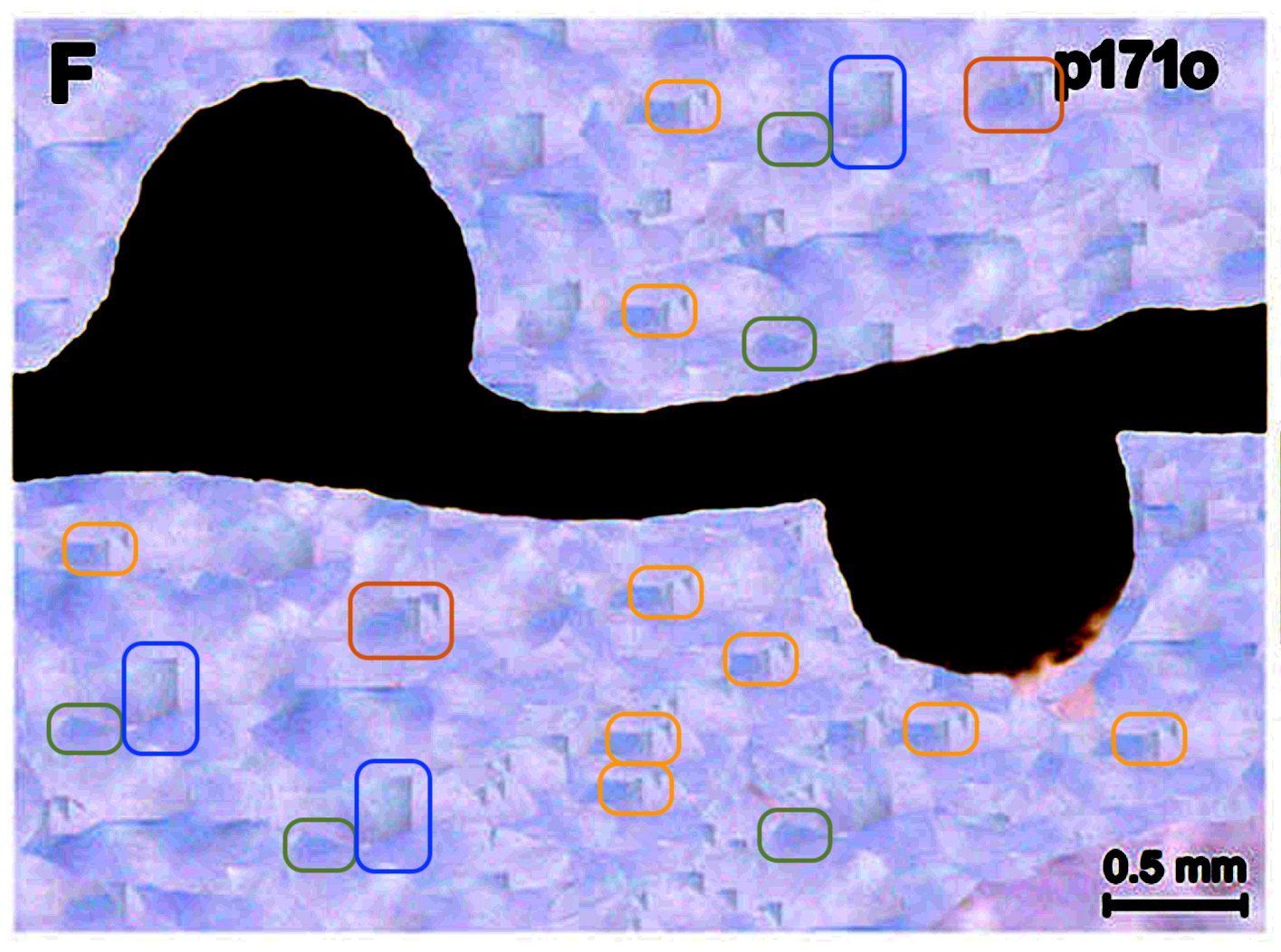
The Photoshop art was spotted by none other than the famous image duplication detective Elisabeth Bik. Look what happened to the soy bean background:
It is not clear what the background beautifications in Photoshop were needed for, maybe to help Figure 1E looks less identical to the Figure 5G? Stacey remains silent on that PubPeer thread.


Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism will be most appreciated!
€5.00



Funny I have to publish stuff on my site to get a reply.
This just in from Michele Kenneth, Associate Vice Chancellor for Research; Research Compliance; Research Integrity Officer at University of Missouri:
“We received your messages and have internal processes to address these matters. “
LikeLike
Here’s an interesting paper vs patent issue.
2013 US Patent US 8,558,056 B2 (original application 2006)
https://patents.google.com/patent/US8558056B2/en
vs
The Plant Cell. 2008 Feb; 20(2): 471–481. doi: 10.1105/tpc.107.056754
Click to access tpc2000471.pdf
The TPC paper reuses 6 figures of the patent, but with some with slightly different legends
LikeLike
Pingback: Help with another? Not on PubPeer yet – For Better Science
A complete ;hatched job…misrepresentation, etc. Everything that the Leonid mentions above has been examined either independently by the University or via the journals themselves and no support for his claims of fraud have been found. The only crime here is to his ego in that he seems unable to accept anything that counters is own skewed view. Oh by the way, I receive no support whatsoever from either soybean industry or commodity groups. If anyone is making false claims, it is Leonid. I suggest, without much of hope of success, that he turn attention to more worthy, positive and productivity undertakings. The ‘get a life’ quote is correct but was not, as indicated above, in direct response to any specific paper or figure. Another misrepresentation.
LikeLike
Dear Gary, I never claimed your university or the journals you publish in would not support you. Quite the opposite. It is actually a misrepresentation from your side. But you cannot seriously do the Giorgio Zauli thing now and say only the official “Science Court” of peer review is permitted to discuss the issues with your figures?
https://forbetterscience.com/2019/08/23/ferrara-protects-rector-zauli-from-prejudiced-media-and-unqualified-public/
LikeLike
Mr Schneider obviously is emotional about my refusal to play along with his tirade. Those issues that have been brought to our attention have been dealt with, either by exoneration via independent investigation, or via posting original figures on pubpeer or, in one case, publishing a correction. These are the legitimate avenues by which one should respond. However, this apparently is not sufficient for Mr. Schneider to desist from his continuing slandering of my reputation. Mistakenly, I took myself off of twitter since this was becoming a distraction. However, I will now pay attention again and comment and defend myself. We continue to do great work and publish high impact papers. I believe, this is the only way to counter such negativism without becoming part of the actual problem.
LikeLike
“The Stacey Lab just spent Two Trillion Dollars on Biology Equipment. We are the biggest and by far the BEST in the World! If Schneider attacks an American lab, we will be sending some of that brand new beautiful equipment his way…and without hesitation!”
LikeLike
Dr. Stacey: to characterize the concerns expressed about your research as a slander of your reputation by Dr. Schneider is to ignore the numerous unaddressed posts on PubPeer authored by numerous readers. That it takes so much effort to encourage you to thoroughly defend your work is perplexing but is the positive result of Dr. Schneider’s persistence.
Couple of examples:
LikeLike
Gary Stacey says his university proclaimed him innocent of everything and a victim of Schneider’s persecution.

Here he “corrected” the Frontiers paper Hossain et al 2019 discussed above. 12 (TWELVE) Figures were REMOVED, 7 figures replaced.
(artwork: Smut Clyde)
Stacey announced it himself:
“Correction has now been published online: DOI: 10.3389/fpls.2019.01692”
This is the “provisionally accepted” correction notice, since the correction is not linked to the article and the doi does not resolve:
I can’t help thinking that Frontiers only agreed to such embarrassingly idiotic correction because they also feel victims of my persecution. Read more here.
LikeLike
Pingback: Society journal Biochemical Reports, ravaged – For Better Science
Pingback: Rick Vierstra probably meant well – For Better Science
LikeLike