Richard Vierstra, born 1951, is a plant science professor at the Washington University in St Louis (WUSTL) in USA who specialises in protein ubiquitylation. He is one of the most eminent researchers in his field, a member of the US National Academy of Sciences (NAS), a former treasurer of the American Society of Plant Biologists (ASPB) and a fellow of the American Association for the Advancement of Science (AAAS).
Vierstra is probably a good and honest scientist, but maybe he is losing it now. It seems he became too trusting towards certain lab members because they were so impressively productive. Worse, Vierstra is stubbornly convinced that the validity of his predicted research results remains unaffected by the data forgery these lab members engaged in to prove him right. He sees conspiracies in the wrong corners.
In January 2022, I wrote to Vierstra with concerns about one of his lab’s papers flagged on PubPeer, one out of several listed there.
Kwang-Hee Lee , Atsushi Minami , Richard S. Marshall , Adam J. Book , Lisa M. Farmer , Joseph M. Walker , Richard D. Vierstra The RPT2 Subunit of the 26S Proteasome Directs Complex Assembly, Histone Dynamics, and Gametophyte and Sporophyte Development in Arabidopsis The Plant Cell (2011) doi: 10.1105/tpc.111.089482

Vierstra immediately replied, and shared an extended figure:

“It does look like something weird happened when formatting this figure for publication in Plant Cell. The extended view of the immunoblot of rpt2a-1 samples with RPN5 antibodies (more fractions than are shown in the Figure) on the right appeared to be displaced to the front of the images to now over hang the anti-RPN5 antibody blot to its left containing rpt2a-3 samples. Once this blot is placed in the back of the gel images, the full gel from the rpt2a-3 samples emerges. There was no attempt to hide any results from the figure. Sorry this error occurred. Not quite sure when it happened (by us in transferring files to Plant Cell or by the Plant Cell publication team after file receipt ) but we have attached the correct Figure 6. When you look at the AI file of the figure, the actual size of the offending image shows how it would have impacted the image to the left if inappropriately brought to the front.“
He also posted the same on PubPeer. Apparently some pictures got shuffled over each others which created a false impression of a gel manipulation. An honest mistake of oversight, nothing serious. The journal’s chief editor Blake Meyers (who in a way got this job basically because of my reporting) announced that “we’ll likely issue a correction.”
There were already some more Vierstra-authored papers on PubPeer back then, but the Vierstra lab seemed overall honest, most importantly the PI seemed dedicated to research integrity.
Also this PubPeer thread initially seemed to be a mix of false alarms and minor issues.
Richard S. Marshall, Zhihua Hua , Sujina Mali , Fionn McLoughlin, Richard D. Vierstra ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors Cell (2019) doi: 10.1016/j.cell.2019.02.009




The above was posted in July 2021, and right away, Vierstra replied on PubPeer and explained everything:
“Thanks for the inquiry related to the Marshall et al. (2019) Cell publication about possible inconsistencies/irregularities about some of its figures. Like you, we the authors are deeply committed to publishing accurate scientific data and have taken your notification seriously. Fortunately, we have found all the original results highlighted by you as possibly being problematic and are now working with Cell to fix any errors. We have also gone over all the other figures to confirm accuracy. From our perspective, it looks like some of the figures highlighted by PubPeer are actually correct and that some have been published in error. Tracing back the errors, it appears that our originally formatted figures were correct in all cases but then some of the panels became altered/substituted by various authors during the iterative improvements/rearrangements of the figures before final publication. Our time-stamps on the succession of figure files allowed us to see when the changes were introduced in error. It appears to be a combined set of errors involving several authors, with none of the authors detecting the problems in the final product. Had we seen these errors, we would have without question fixed them promptly. Importantly, these placement errors in no way altered the outcomes nor our interpretations in the Cell publication, and can easily be rectified by replacing the panels published in error. We hope to have the corrections complete in the near future.
Sincerely, Richard D. Vierstra Richard S. Marshall“
Vierstra and Marshall then shared on PubPeer raw data for all the criticised figures.
In March 2022, an Erratum was issued, fixing “errors in Figures 7 and S1,” and reassuring that “Both errors did not affect the results in the paper nor the interpretation of the data“. The plant science community was thankful to Vierstra’s lab and the editors of Cell and their publisher colleagues at Elsevier who have nothing but ethics and integrity on their minds.
And then Elisabeth Bik had another look. Simply put, it seems what Vierstra and Marshall submitted as “raw data” was actually forged.
Bik’s conclusion:
“I am not sure about the author-provided originals showing the yeast assays. Both the published figures as well as the provided originals look very regular, without the small irregularities in shape and position normally observed in these assays. They look like nearly-perfectly white circles on a perfectly black background.“


Funny how you now suddenly see the yeast traces in negative samples which Acronicta ovata found completely blank, but now the previously seen dots and other irregularities disappeared without a trace. Same here:

And then Bik found also this:



A bunch of very, very fraudulently fake gels. Combined with the earlier suspicion that the yeast assays are also simply fabricated, this all does affect the results in the paper as well as the interpretation of the data.
Bik found even more, apparently our charming friends at Cell and Elsevier already stealthily corrected that Vierstra paper soon after it was published, sneakily altering the allegedly permanent scientific record. The Elsevier’s version is different from the PubMed version deposited right after the paper was first published. Some fudged figures were stealthily replaced by Cell and Elsevier, most likely upon authors’s request.

Vierstra wasn’t at a loss for words. On PubPeer, he insisted that “Cell has not silently updated the figures” (Then who did? The Tooth Fairy?), and that “Despite the irregularities, I am not aware that any of the conclusions from the paper have been compromised.”
Don’t expect Cell to retract the paper now, that was exactly why they issued the stealth correction and the recent the Erratum so quickly before anyone notices something untoward. Especially since Vierstra insists that conclusions are not affected by much of his paper’s data being forged.
Vierstra also mentioned on PubPeer:
“The lead author, Dr. RIchard Marshall is no longer in the lab has not yet responded to my requests for comment related to several of your recent concerns.“
Indeed, there were many problems with Marshall’s productivity which Vierstra has been trying to fix in the last months.
This Molecular Cell paper (with same Elsevier’s Cell Press) was corrected in May 2021:
Richard S. Marshall , Faqiang Li , David C. Gemperline , Adam J. Book , Richard D. Vierstra Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis Molecular Cell (2015) doi: 10.1016/j.molcel.2015.04.023
The Correction notice stated:
“We, Richard S. Marshall and Richard D. Vierstra, recently identified an error in this 2015 Molecular Cell paper. In Figure 3C, a pair of confocal images (PAG1-GFP N +ConA and RPN5a-GFP N +ConA) were inadvertently duplicated within the figure, and a pair of confocal images (PAG1-GFP +N ConA and RPN5a-GFP +N ConA) were incorrectly duplicated from Figure 1C.“
What Bik visualised now, does not look like inadvertent errors, but like something very dishonest.

Or how about this corrected paper by Marshall:
Richard S. Marshall, David C. Gemperline, Fionn McLoughlin , Adam J. Book , Kay Hofmann, Richard D. Vierstra An evolutionarily distinct chaperone promotes 20S proteasome α-ring assembly in plants Journal of Cell Science (2020) doi: 10.1242/jcs.249862

Vierstra and Marshall replied on PubPeer in December 2021: “we affirm that the conclusions of the paper are correct“.
The learned society Company of Biologists and their Journal of Cell Sciences swiftly issued a Correction in January 2022:
“In Fig. S2A, two bimolecular fluorescence microscopy (BiFC) confocal microscope images were inadvertently duplicated in the bottom panel. These panels, which both contain only control background fluorescence, were not used to infer any positive interactions and the error does not affect the results or conclusions of the paper.”

It is a pity that this Company of Biologists wasn’t interested in checking this paper properly. Well, Bik did it for them, gratis, and found yet another problematic figure. Will they now retract the forgery, with its unaffected conclusions? They should, or do they want to be like Elsevier?
Vierstra, once so eloquent and ready to post raw data, remained silent. He probably is currently negotiating with Elsevier a correction notice to this Cell Press paper to declare all conclusions unaffected:
Richard S. Marshall, Richard D. Vierstra A trio of ubiquitin ligases sequentially drives ubiquitylation and autophagic degradation of dysfunctional yeast proteasomes Cell Reports (2022) doi: 10.1016/j.celrep.2022.110535

The referenced preprint Marshall & Vierstra 2021 was posted on bioRXiv on 13 May 2021, the manuscript submitted to Cell Reports on 9 June 2021. Vierstra explained on PubPeer (typos his):
“During our own extensive review of the work along with the detailed peer review process by Cell Reports over the past year, we have adjusted interpretation of several parts of the study, corrected some mistakes, and added new experiments suggested by peer review or decided by us as useful to the work. The errors you note were found by us and promptly corrected. We have contacted BoRxiv and asked them to deleted their outdated version of the paper on their postings. Sorry for the confusion. Please refer to the Cell Reports paper for all further interest in this study.“
Really? Extensive review? Adjusted some mistakes? Bik found this, in the Cell reports paper, mind you:

And the same kind of fake gels we met before:


Plus this, inappropriate image reuse from an earlier Marshall et al paper:
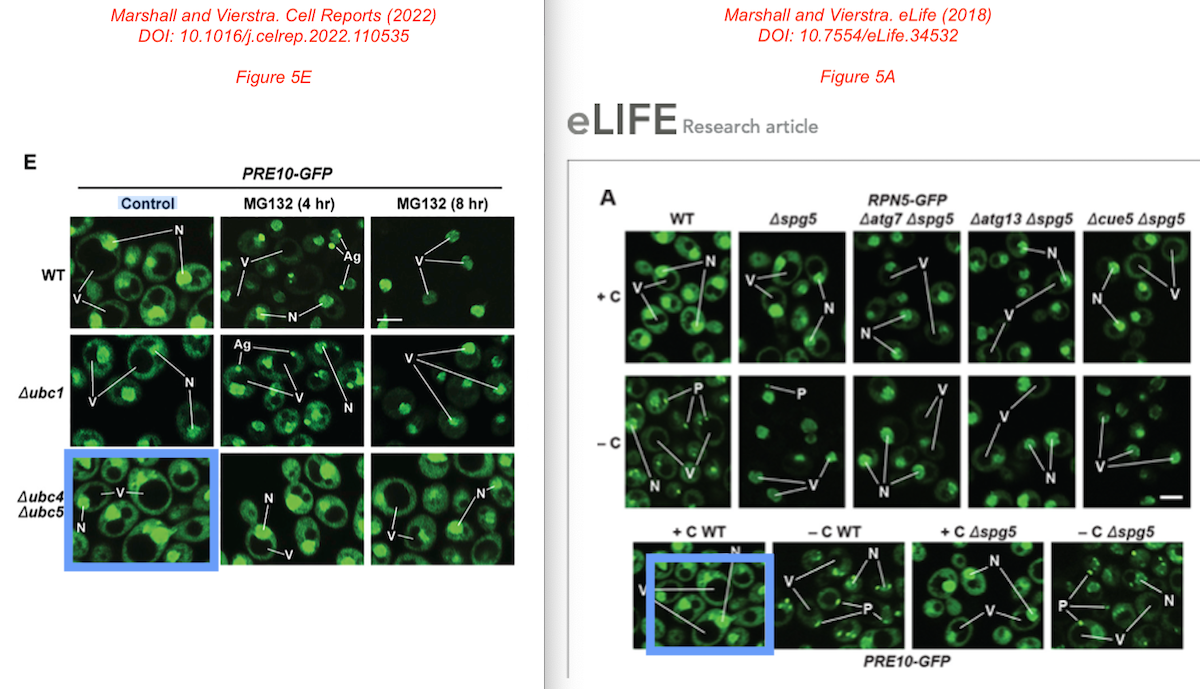

No comments from Vierstra anymore. Conclusions still unaffected?
Now, that eLife paper is problematic in itself:
Richard S Marshall, Richard D Vierstra Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation eLife (2018) doi: 10.7554/elife.34532

PubPeer user Huangyuannia tibetana thought in November 2021 that in Figure 4, “Panel D The lower GFP bands look the same including the backgrounds.”
But Marshall and Vierstra stated on PubPeer:
“We agree that the bottom portions of these blots do look somewhat similar, but we think a close inspection of the raw data will show that they are not duplicates.”
They also provided the “Original TIF file” image of the gel.
But then someone spotted a problem in yet another figure panel.

Also here, Vierstra shared in January 2022 what is supposed to be raw data and declared:
“The Figure 3F in question was generated by the first author. From comparisons of the published figure to the original x-ray film images of the two blots (see appended figure), we conclude that the figure is correct as is despite some apparent similarities in band appearances for free GFP.“
The case was presumably closed. But since that time, Bik exposed new problems with Marshall & Vierstra papers, and one wonders how real those original gel images really were.
Is Vierstra’s explanation for this Marshall-coauthored paper still trustworthy?
Nicholas P. Gladman , Richard S. Marshall , Kwang-Hee Lee , Richard D. Vierstra The Proteasome Stress Regulon Is Controlled by a Pair of NAC Transcription Factors in Arabidopsis The Plant Cell (2016) doi: 10.1105/tpc.15.01022

A simple explanation would be that someone digitally inserted those gel bands where there were none. But not for Vierstra, who in January 2022 again shared on PubPeer “raw data” and declared:
“The Figure 1 panel B in question was generated by the first author. From comparisons of the published figure to the original blot (see appended figure), we concluded that that figure is correct as is, and that no manipulation of the figure occurred.”
There were more Marshall & Vierstra papers flagged on PubPeer, some were corrected, some were not because the “raw data” proved the allegations unfounded.
Richard S. Marshall, Fionn McLoughlin , Richard D. Vierstra Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone Cell Reports (2016) doi: 10.1016/j.celrep.2016.07.015

Vierstra only admitted the H3 blots to be duplicated, and issued in March 2022 a Correction for just that (“The change does not impact the conclusions of the study“). For the GFP bands, Vierstra stated:
“We do however feel that the additional issue highlighted by PubPeer for a separate region of Figure S3 is not justified. Analysis of the raw data for the two panels where there is concern about the lower free GFP bands (the GFP-APE1 and PEX14-GFP blots) being the same indicates to us that they are not.“
Also in the next case, Vierstra only admitted full blot duplications, never those of a single gel band. That’s because the former can be theoretically explained by an honest mistake of oversight, but the latter cannot.
David C. Gemperline, Richard S. Marshall , Kwang-Hee Lee , Qingzhen Zhao , Weiming Hu , Fionn McLoughlin , Mark Scalf , Lloyd M. Smith , Richard D. Vierstra Proteomic analysis of affinity-purified 26S proteasomes identifies a suite of assembly chaperones in Arabidopsis Journal of Biological Chemistry (2019) doi: 10.1074/jbc.ra119.010219



In December 2021, Marshall & Vierstra shared “raw data” and announced on PubPeer:
“We have now researched the outlined concerns and associated raw data and, although we affirm that the conclusions of the paper are correct, we did confirm two of the errors in Figure 2 and 10C highlighted above that involved the incorrect placement of immunoblots during construction of Figures containing glycerol gradients. […]
We do however feel that the additional issue highlighted by PubPeer for Figure 3B is not justified. Analysis of the raw data for the three panels highlighted in Figure 3, where there is concern about the similar shape of some bands, confirms that these are indeed separate blots. […]
The errors will be corrected in an upcoming issue of the Journal of Biological Chemistry in the near future.“

To me, Vierstra wrote this in an email:
“First off, I want to emphasize that I, like almost all of us, want strong mechanisms to enforce ethical publishing PubPeer, if used ethically, can be one mechanism. And yes Richard Marshall has been sloppy in his figure preparations, which is distressing in-and-of-itseff. We have had a number of discussion in my lab on this issue. Fortunately, at this point, we are unaware that any of his conclusions are wrong.
But…. you should be aware that it appears that PubPeer is being used by someone (or someones) to get at Richard personally. I am not at liberty to speak about this legally at this point, but I know of at least three other anonymous avenues that this person(s) has used to attacked him out side of PubPeer comments. Its clear that this person(s) wants to cripple him and his scientific career. This person(s) also knows Richard personally based on the mechanism(s) used that only a person with intimate knowledge of him would know. Richard is such a mild mannered guy that I don’t understand this hostility. I feel that PubPeer is now being used as only one method to attack him. Unfortunately, these approaches may have worked. Richard left my lab in February when his visa expired and he is no longer responding to any emails from anyone. We have no idea where he is at the point and have no way to reach out to him.[…]
So for you as “manager” of PubPeer, what is the mechanisms to control its ethical behavior? Right now this PubPeer commentor(s) is behaving as the judge and jury under the cloak of anonymity. I feel that these continuous comments(s) may be pushing the boundaries, which should also be a concern for PubPeer.
I should add that we sent all the relevant information requested by Cell on his 2019 paper related to PubPeer comments a number of months ago. The Cell editors reviewed the work and the original images, and only requested that we submit an Erratum that fixed a duplicated gel. This we did months ago. You might not like how Cell handles questioned data, but we followed their protocols are requested.“
I wrote back to Vierstra:
- “Dr Bik has no conflicts of interests, and her expertise is beyond doubt. The recent pubpeer posts are all by her.
- Several articles flagged by Dr Bik do not feature Mr Marshall as coauthor. I am not sure I understand why you think those posts have anything to do with him. Like this one, with allegation of raw data forgery. https://pubpeer.com/publications/D44C032AB3BFA894253215F77F695F
- I have no affiliation with PubPeer, I do not comment there for some years already, and I am probably the last person whose input the PubPeer operators are interested in.”
I also suggested Vierstra requests a research misconduct investigation. He did not reply anymore. This was the paper I mentioned, it’s over 20 years old, done long before Marshall joined Vierstra’s lab.
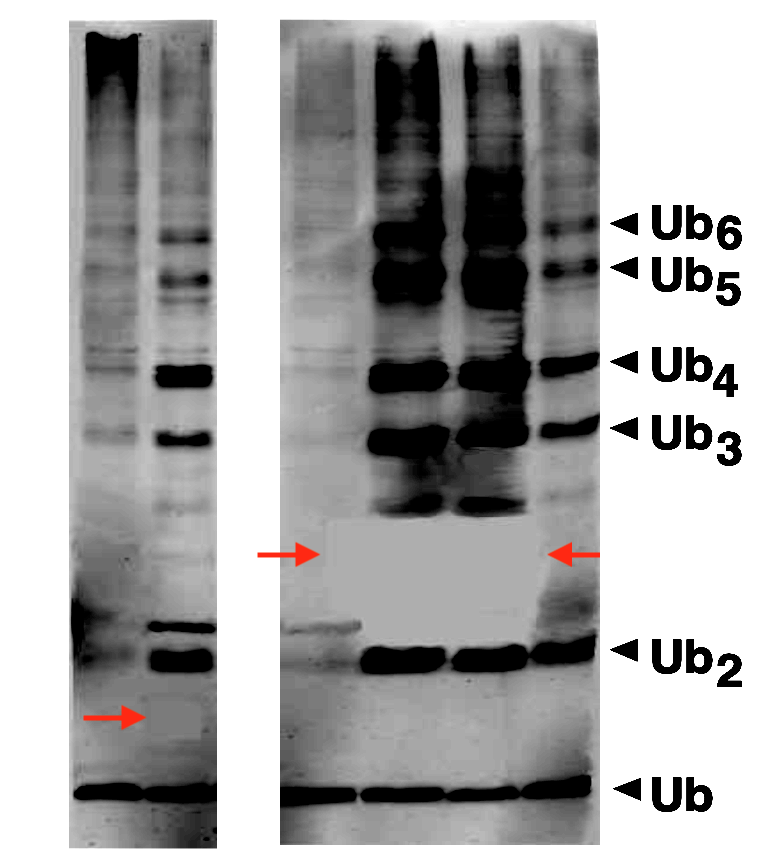
Jed H. Doelling, Ning Yan , Jasmina Kurepa , Joseph Walker , Richard D. Vierstra The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana The Plant journal (2001) doi: 10.1046/j.1365-313x.2001.01106.x

In this Fig 3B, Bik noted in May 2022 that “A remarkably blank area can be observed in the right panel, between the Ub2 and Ub3 bands.” Vierstra disagreed and shared raw data to prove her wrong.
Bik found even more digital forgeries in the “raw data”:
“The higher resolution photo of Figure 3B you provided here confirms the completely blank areas, which have a remarkably sharp rectangular shape, without the specks and graininess observed in the other parts of the photo. In fact, a third blank area with a sharp rectangular corner is visible, just under the UB3 band in the C317S lane.“
Vierstra was not prepared to admit defeat:
“I have no idea where these boxes come from. The figure was constructed over 22 years ago and the lead author responsible for this figures is no longer in science. Fortunately, the weird issue was not near the main parts of the immunoblot generated with anti-ubiquitin antibodies.“
Basically, so what the published figure and the “raw data” are fake, conclusions still remain unaffected.
At some point Vierstra should consider the possibility that certain lab members of his were simply serving him fake data to prove his preconceived theories, which he converted into papers in high-ranked journals and research grants. It does not matter anymore if Vierstra’s theories were right or wrong, the only unaffected conclusion is that fraudulent science must be retracted.
And again, it was not just Marshall who created problems.
Xiao Huang , Chunyan Zheng , Fen Liu , Chao Yang , Ping Zheng , Xing Lu , Jiang Tian , Taijoon Chung , Marisa S. Otegui , Shi Xiao , Caiji Gao , Richard D. Vierstra, Faqiang Li Genetic Analyses of the Arabidopsis ATG1 Kinase Complex Reveal Both Kinase-Dependent and Independent Autophagic Routes during Fixed-Carbon Starvation The Plant Cell (2019) doi: 10.1105/tpc.19.00066

Vierstra swiftly commented on PubPeer:
“Dr. Faqiang Li, the corresponding author is going to handle this inadvertent duplication of the seedling photos.“
How does he know this duplication was inadvertent? Because this is also what Faqiang Li, now professor at the South China Agricultural University, but until 2015 postdoc in Vierstra’s lab, says:
“Thank your very much for identifying our inadvertent duplications of the images in Figure 4A! This was indeed an unintentional duplication. I will contact the editor to see how we can get that corrected.“
Vierstra decided not to invite Li to explain these inadvertent duplications:
Faqiang Li , Taijoon Chung, Richard D. Vierstra AUTOPHAGY-RELATED11 Plays a Critical Role in General Autophagy- and Senescence-Induced Mitophagy in Arabidopsis The Plant Cell (2014) doi: 10.1105/tpc.113.120014

Red boxes: The NY-ATG11+CY-ATG101 panel in Figure 3C appears to show the same specimen as the NY-ATG101+CY-ATG11 panel in Figure S7C, albeit shown at different magnifications.“

It is strange Vierstra insists that Li’s duplications are inadvertent, what with the Marshall story and this correction the WUSTL professor already hat to issue for Li:
Anongpat Suttangkakul , Faqiang Li, Taijoon Chung, Richard D. Vierstra The ATG1/ATG13 Protein Kinase Complex Is Both a Regulator and a Target of Autophagic Recycling in Arabidopsis The Plant Cell (2011) doi: 10.1105/tpc.111.090993


The December 2021 Correction proudly assured:
“The results and conclusions of this work are unaffected by these corrections.”

In an ASPB interview from 2020, Vierstra gave this advice to early career researchers in plant sciences:
“First off, be dedicated to your work and the quality of your science.
There are really no shortcuts to having a successful career.”
Time to live by your own advice, draw the correct unaffected conclusions and retract your problematic papers, Professor Vierstra.
The article was corrected on 14.06.2022 to correctly describe the current affiliation of Faqiang Li.
Update 22.06.2022
Now I am not that sure anymore that Rick Vierstra meant well.
Remember him telling me regarding Richard Marshall:
“Richard left my lab in February when his visa expired and he is no longer responding to any emails from anyone. We have no idea where he is at the point and have no way to reach out to him.“
Now I received this email:
“I’m Richard Marshall, Rick Vierstra knows very well where I am. I’m still in st Louis, at my apartment on Skinker Blvd.“
Vierstra did not reply when confronted with this message. But he also chose not to deny some much heavier accusations, raised in this Inside Higher Education article from 2018, authored by his former lab member Nabil Elrouby.
By now three former Vierstra lab members confirmed to me that the professor X, professor Y and Professor Z in this article are all Vierstra. And Vierstra did not deny this, despite my repeated asking. This is what Elrouby’s article says:
“Take the example of Professor X. In that professor’s lab, several graduate students and postdocs (past and present) needed psychiatric assistance and prescription drugs to be able to deal with his pressure and management style. Students who spent six or seven years on their Ph.D. projects, and published work, with promising results that could contribute to other publications, were told that they “will not be able to graduate” and that they “may have to master out” — in other words, to leave with only a master’s degree instead of a Ph.D. Professor X designed the graduate projects, and as a student in his lab you could not deviate from the experimental plan that he declared for you.
But when results were not favorable, he did not claim responsibility but instead blamed the students. He cut off funding, yet still demanded his grad students to work full-time in the lab for years. He used reference letters as bargaining cards to keep his grad students and postdocs from leaving the lab — even at times when they received no financial support — to get them to do the experiments that would satisfy his unreasonable requests. “
Now it kind of puts the problems with the papers by Richard Marshall and Faqiang Li into a new light.
“Consider another example. Professor Y visited a research institute abroad and met with different people who had similar research interests. Through a discussion with Bob (a pseudonym), a senior postdoc, Professor Y got to know about Bob’s findings while offering no details about his own. A year later, Professor Y met Bob’s postdoctoral adviser in a scientific meeting and inquired about the status of Bob’s project. He learned that Bob was writing a paper for a top-tier journal. Professor Y suggested that he, too, was writing a paper and suggested that he and Bob would benefit from “coordinated” publication of the results by submitting the two manuscripts simultaneously.
But Bob had already finished writing his manuscript while Professor Y needed a few more weeks to finalize his data and write it up. He asked Bob for his data set to compare it with his and promised to reciprocate. Months later, and after several email inquiries in which Bob asked whether Professor Y would send his data as promised and whether it was ready for submission, Bob and his adviser were shocked to hear that Professor Y had already single-handedly submitted his manuscript.
When questioned about it, Professor Y blamed his submission on his graduate student (and first author), although the grad student had no choice about when or if to submit the manuscript. In fact, Professor Y had disregarded a written agreement between Bob and himself that they would not scoop each other. “
That is probably why Vierstra is so successful and his science is so reproducible despite evidence of data manipulations? Because he runs off with other lab#s results to publsih them first? And when the losers finally publish their findings in a lowly “specialised” journal, Vierstra uses it as proof of reproducibility of his papers?

“A third story involves Professor Z. Professor Z met Mike (also a pseudonym) in a scientific meeting. Mike presented new data that he came across through a carefully designed experimental plan. When Professor Z could not obtain enough details about such unpublished data, he accused Mike of concealing some, which Mike assumed to be unnecessary details that would appear only with the publication of his work.
But Professor Z wanted the data so badly that he offered Mike a job in his lab. Mike was hoping for a faculty position, but his funds were running out. He accepted Professor Z’s offer, with a verbal agreement that he would bring his projects with him and would use his short tenure at Professor Z’s lab to complete and publish several of those projects. The agreement was supposed to serve both Mike and Professor Z well: Mike would get his half-done projects published, and Professor Z would get his name on them.
As soon as Mike joined the lab, however, Professor Z inquired about the data Mike had presented in the meeting. When he learned that the data was already written for publication, Professor Z became extremely agitated and suggested that all the material Mike generated in the past was now his (Professor Z’s) material because it was physically present at his university. Soon after, he asked Mike to stop working on his projects and to help a graduate student in the lab set up an assay that Mike had expertise with.
Although that would violate their agreement, greatly derail his progress and hamper his efforts to publish his work and apply for faculty positions as planned, Mike agreed to do so since Professor Z was paying his salary, after all. But as soon as Mike set up the assay, Professor Z declared that he’d run out of money and that Mike had to leave the lab. When the work that involved that assay was about to be published, Professor Z insisted on doing that himself without including Mike’s name in the author list. Mike contested the issue repeatedly with Professor Z, putting together evidence that illustrated his contributions to the work.
But it became clear that Professor Z had no intention of having Mike as a co-author on that paper.
Mike raised the issue with the publishing journal, but his evidence was outweighed by a letter from Professor Z suggesting that, because he’d been a professor for more than 30 years, he was able to judge who should be author. Moreover, he claimed that Mike was just a zealous young scientist who saw the benefits of publication and “unjustifiably” wanted to be an author on that paper.
Mike also raised the issue with Professor Z’s university officials, who suggested that since Mike wasn’t questioning the integrity of the findings themselves, the decision of authorship was left to Professor Z. To Mike, it was clear that Professor Z’s hopes were that he would host him for a short period of time, get hold of the material that he generated as a postdoc in his previous institute and then let him go.”
Maybe this is why Vierstra moved his lab to WUSTL in 2015, abandoning the University of Wisconsin where he was a faculty member since 1984. Maybe they had enough of his antics.
And those who don’t know Vierstra close enough, adore him as a charming science genius:
“Outside a professional setting, Professors X, Y and Z are gregarious, talkative and charming. They can recruit starting grad students and postdocs who do not know the inside stories. Once those students and postdocs are in their graduate programs or research projects for a couple of years, it is difficult for them to leave without a degree or a paper. So, they are stuck. Previous grad students and postdocs couldn’t communicate this information about Professors X, Y and Z, as they feared retaliation. After all, their careers depended on a letter from their PI.”
I thank all my donors for supporting my journalism. You can be one of them!
Make a one-time donation:
I thank all my donors for supporting my journalism. You can be one of them!
Make a monthly donation:
Choose an amount
Or enter a custom amount
Your contribution is appreciated.
Your contribution is appreciated.
DonateDonate monthly

“There are really no shortcuts to having a successful career.” Lol! I think he mispoke: should have said “you must short-cut as necessary to get papers published in high impact factor journals so that you satisfy the universities requirements for a successful career– getting grants with a lot of overhead.” In short, pull a Sabatini.
LikeLiked by 1 person
I do not understand Vierstra.
There’s nothing shameful to retract papers when one acknowledges there are manipulations, quite the contrary actually. Vierstra’s stubbornness in wanting to defend the indefensible is worrying.
Here we are facing a well known scientist, aged 71, with little to gain in saving those papers and with a 50 years long career at stake. Again, I cannot understand.
LikeLike
Dr. Vierstra seems to be blaming other researchers when these problems go back over 20 years. He should look at the common denominator (himself) for answers as to why this keeps happening.
LikeLike
Not strictly related to the problems highlighted by this report:
The crazy circus of academic congresses and conferences is back on track. For instance, on 23-27 October the 13th International Congress on Plant Molecular Biology (IPMB 2022) will take place in Cairns, Queensland, Australia. Among the plenary speakers there’s also Richard Vierstra. Of note, another plant scientist with a serious PubPeer record, Julia Santiago, will be invited to chair the “Structural Biology” session. Thousands (!) of people will cross oceans and continents to present… powerpoint slides.
Really?
Are the participants aware of the massive carbon footprint genarated by this nonsense?
Is this just a way to further milk out taxpayers money to leech travel costs for holidays?
Could academia please ban forever global congresses in exotic and remote places, and organize instead meetings locally? And eventually with a very few people (two-three individuals maximum!) invited from overseas?
LikeLike
Ooh! At this unmissable IPMB 2022 congress also Georgia Drakakaki will be invited!
Lovely!
LikeLike
You got it all wrong. All these scientists don’t come to simply listen to Julia Santiago’s talk, they come to express support to her and other cheaters and to discuss ways to defraud the public with fake science.
LikeLike
Two corrections:
“Faqiang Li, a senior scientist based not somewhere in China, but in Vierstra’s lab in WUSTL”
Faqiang Li did not join Rick Vierstra in moving to WUSTL in 2015, and did move to China. A 2021 paper lists his current institution as South China Agricultural University, Guangzhou, China.
“Marshall is still officially listed as a member of Vierstra’s lab”
The link here is to Rick Vierstra’s lab website from the University of Wisconsin, which hasn’t been updated since he left the university for WUSTL in 2015.
LikeLike
Thanks for the corrections! Will fix the article.
LikeLike
That eager young mind of Richard Marshall, who early understood one should cheat hard to succeed in science, was expected at the PPSTASIS 2022 Congress in Madrid on 21-23 September as invited speaker: http://plantproteostasis.com/speakers/
However, the day-by-day program released hours ago, doesn’t include him nor another invited speaker with a serious PubPeer record: Karin Schumacher.
For a sec I thought this double defection was due to research integrity concerns by the organizing committee, but then I realized Vicente Rubio is among them.
LikeLike
The “Inside Higher Education” article from 2018 by Nabil Elrouby is mind-blowing. A must read.
Vierstra is 71. It’s time he ritires, for good.
LikeLike
LikeLike
LikeLike
We can only hope that, after a complete and thorough investigation, the school throws this apparent sociopath under the bus.
The sociopath is, of course, Vierstra.
LikeLiked by 2 people
@Regret, if you look at the screenshot on Twitter compared to the same post on pubpeer now, you can see that Professor Richard Vierstra has deleted the sentence:
“Major complications were the inadequate response by the first author, who departed the laboratory in February, 2022, his poor data archiving, which has left me with trying to inventory the reams of photos and gel images associated with the project, his ongoing personal issues, and additional concerns that emerged during the review process.”
https://pubpeer.com/publications/0340E8A16B24B210544B9910D7C63E
LikeLike
Yes, I saw. That’s the reason I took a screenshot. Perhaps the first author will get some mileage out of it.
LikeLike
Retraction for the disastrous eLife paper Marshall & Vierstra 2018:
https://elifesciences.org/articles/82988
LikeLike
I, as the last author, was responsible for ensuring scientific integrity for this publication but clearly failed as a ‘gatekeeper’ in this case. The corresponding author Richard D. Vierstra agrees to this retraction. The following author was also contacted but he did not provided comments nor explanations for the errors: Richard S. Marshall.”
did not provided
Dr Vierstra, learn English.
LikeLike
“As the corresponding author I retract the eLife paper cited above based on unexplained irregularities associated with a collection of western blots and inappropriate placement of several confocal fluorescence micrographs generated by the first author Dr. Richard S. Marshall who was responsible for all the experiments and the first preparation and final drafts of the figures.”
Wow. Vierstra threw Marshall right under the bus….Hope Marshall doesn’t have moths to feed. I think Vierstra needs to move away from the US to Sicily, whose research ethics appear to be more consistent with his.
LikeLike
I just fed 2kg of Mejoul dates and a kilo of delicious pistachios to the moths, plus other foodstuffs, don’t get me started.
LikeLike
Got to keep moths fed or they start eating your clothes. Hence, the reason I have holes in my clothes, because as a failed scientist I cant keep my moth colonies fed. When you’re a former perma-postdoc like me, this is what happens…moth feeding failure (mff) is a marker for penury.
I wonder if the great Sabatini is keeping his moths fed.
LikeLike
Food moths and wool moths are different species. Lavender helps for latter, nothing at all helps for former, the commercial traps are useless.
I will listen to Prof Raoult’s science and start to douse everything with ivermectin and chloroquine now, that will show them.
LikeLike
look who just get funded by NIH https://reporter.nih.gov/search/Q27knwROxUa3mXM1WZRH1g/project-details/10366935
LikeLike
Hi Richard, how do you feel about the text of this retraction notice? https://elifesciences.org/articles/82988
LikeLike
Vierstra just awarded an NIH R01 grant with all the data Richard Marshall generated in the past 10 years at the beginning of Aug, the same time WashU started an official investigation into the integrity of the research of Vierstra Lab. I guess the rule of presumed innocence until proven guilty really worked here…..
LikeLike
When “mistakes” are found time after time and there is a common factor, your collective assumption is this is a great career with a few problems at the end.
Another conclusion might be that this person long ago discovered that fake results won’t be doubted, won’t be caught, but will get you on the gravy train of grants, international conferences and back slapping.
Go back 40 years and you may well find the same data fixes.
LikeLike