The following guest post by an image integrity sleuth who comments on PubPeer as “Aneurus Inconstans” is about a star of plant science research, Natasha Raikhel: emeritus professor at University of California Riverside and member of the US National Academy of Science. A number of her papers were criticised on PubPeer for data irregularities. Raikhel announced to investigate and to correct the literature in every single case, but the outcome the public gets to see does not really follow her announcement:
“You can be assured that neither I nor any of my colleagues […] would tolerate any data manipulation.“
Raikhel was born in 1947 in Leningrad (now St Petersburg), and left the Soviet Union for USA in 1979, having just survived a plane crash. In 1986, she became faculty member at the Michigan State University. In 2001, while recovering from breast cancer, Raikhel moved to UC Riverside, where she founded the Center for Plant Cell Biology, and in 2016, she retired aged almost 70.
The problems in some of her publications are most probably not Raikhel’s fault, it is just very unfortunate that in almost all cases her junior co-authors, now all senior scientists themselves, fail to live up to the standards of good science their mentor has set. Which is a pity, after the Olivier Voinnet affair the field of plant sciences became particularly sensitive to research misconduct. In the previous data manipulation affair uncovered by Aneurus Inconstans, blame accusations flew in all directions:
Científicos al borde de un ataque de nervios
Dead men don’t talk. A dead colleague, especially a foreigner, is a perfect scapegoat to blame for fake data in your papers. And in your own PhD thesis.
Caught in a SNARE
By Aneurus inconstans (with LS)
In eukaryotes, transport among organelles is mediated by vesicles that bud from a donor membrane and fuse with another. This fusion requires the correct pairing of specific cognate SNARE proteins on the vesicle surface and on the acceptor membrane. SNARE proteins contain a conserved membrane-proximal heptad repeat sequence known as the SNARE-motif. The trans-assembly of motifs into a four-helix bundle drives the fusion between lipid bilayers. Complexes of different SNAREs will target different membranes for delivering different cargoes. The understanding of SNAREs mechanism of action was one of the major accomplishments made by James Rothman, 2013 Nobel Prize winner. Plants are green, don’t move nor they scream, and look very different from animals. However, they are eukaryotes too, thus what stated above holds true.
The Russian-born, adopted-American plant scientist Natasha Raikhel, who retired in 2016, is a NAS member and a bigwig in the field of protein transport in plants. Back since mid ’90s she has significantly contributed on the study of plant SNAREs, at Michigan State University first and at UC Riverside later. Sanderfoot et al. Mol Biol Cell 2001 comes from Raikhel’s most productive years, it provides a combinatorial analysis on the interactions among several Arabidopsis thaliana SNAREs via immunoprecipitation (IP) by using the microsomal fraction extracted from root tissues and affinity-purified antibodies covalently linked to protein-A-Sapharose beads. The study concludes that at least five different SNARE complexes are working along the transport route between the Golgi apparatus and the vacuole. This is a very well cited paper on plant SNAREs with 255 citations as of today (Google Scholar), and it keeps being cited well because few people use the PubPeer plug-in, as it turned out in January this year to be very problematic.
Anton A. Sanderfoot, Valya Kovaleva , Diane C. Bassham, Natasha V. Raikhel Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell Molecular Biology of the Cell (2001) doi: 10.1091/mbc.12.12.3733

The duplicated panels are all showing negative results, as the interaction between those pairs of SNAREs is shown not to occur (third lane empty). These panels corroborate the take-home message that distinct complexes exist, as not all SNAREs interact with one another. However, are the positive interactions detected reliable? We’ll come to this point soon.

First author is Anton Sanderfoot, now associate professor at the University of Wisconsin–La Crosse, third author is Diane Bassham, now professor at the Iowa State University, while second author Valya Kovaleva is no longer in academia. Many duplications spread in just two figures, with flips, stretches, squeezes and contrast changes. One may wonder how all this chaos could occur by mistake. Authors couldn’t retrieve the raw data, after all the paper celebrated its 20th birthday this year. The reputable journal MBoC and its Editor-in-Chief started a commendable investigation, which cannot be taken for granted on such occasions due to the time elapsed, however, the outcome is odd. No expression of concern by the journal, but an inaccurate and short Correction was issued instead in September 2021. It reads:
“The authors of “Interactions between Syntaxins Identify at Least Five SNARE Complexes within the Golgi/Prevacuolar System of the Arabidopsis Cell” (Mol. Biol. Cell [2001] 12, 3733–3743; 10.1091/mbc.12.12.3733) wish to note errors in their paper.
Several mistakes in the assembly of Figure 4 resulted in the duplication of bands that were actually from Figure 5. Unfortunately, due to the length of time since publication, retirement of the corresponding author and one other author (N.V.R. and V.K.) and all of the authors no longer at the site of the work since the time of publication, the primary data are no longer available to support correction of the figure. The authors stand by the conclusions of the paper and note that multiple other independent labs have since confirmed their findings using independent reagents or mass spectroscopy (Yano et al., 2003; Niiharma et al., 2005; Ebine et al., 2008; Kato et al., 2010; El Kasmi et al., 2013; Fujiwara et al., 2014; Takemoto et al., 2018). The authors regret the errors and apologize to the community.“

No information on which panels were duplicated, no mention that duplications occurred also within Figure 4. Authors also state that duplications in Figure 4 were from Figure 5, but how do they know as they no longer have the raw data and cannot check? Beside the misspelling of a citation in the text (Niihama et al. 2005), it all reads as if written in a five-minutes coffee break. Is it true that those thirteen interactions reported in Sanderfoot et al. 2001 were confirmed by those studies cited by the authors? Not all of them, actually. Six were confirmed (SYP21-VTI11, SYP21-SYP51, SYP22-SYP51, SYP22-SYP52, SYP51-VTI11, SYP51-SYP61 by Yano et al. 2003, Kato et al. 2010 and Fujiwara et al. 2014), four weren’t investigated (SYP21-SYP52, SYP51-VTI12, SYP52-SYP61, SYP61-VTI12), and three were not confirmed (SYP41-VTI12, SYP41-SYP42, SYP41-SYP61 by Kato et al. 2010 and Fujiwara et al. 2014). Four out of seven cited papers do not deal at all with those interactions found by Sanderfoot et al. 2001, maybe the authors thought it was good to add a few more references in to impress the editorial board?
Not a good look for the journal, but it can be worse!
Meaningless and pseudoscientific potatoes
How to cook potato data. A recipe from Poland.
It is a very strange correction, especially in view of Raikel’s email (to LS), where she declared in January 2021:
“You can be assured that neither I nor any of my colleagues are aware or would tolerate any data manipulation. I and the people in my lab were always very careful and 90% of the time every blot was run by two different people and independently confirmed.“

And yet they did tolerate the data manipulation. Maybe for this reason, as Raikhel explained regarding the Sanderfoot et al 2001 study:
“The accusation for the work that was done in Michigan State University is even more difficult to assess. I left the University a long time ago and the first Author of the paper is working on a completely different topic and does not have any records left. After I left, all the lab books were disposed of.“
When such a bad case as Sanderfoot et al. 2001 is found, it’s usually worth investigating the whole publication record of the lab involved, and indeed eight more papers from the Raikhel’s lab have issues. For instance Rosado et al. Plant Cell 2010, for which another strange correction was issued this year and already reported in Schneider Shorts of 23.04.2021.
Abel Rosado, EunJu Sohn, Georgia Drakakaki, Songqin Pan, Alexandra Swidergal, Yuqing Xiong, Byung-Ho Kang, Ray A Bressan, Natasha V Raikhel Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis The Plant Cell (2010) doi: 10.1105/tpc.109.068320
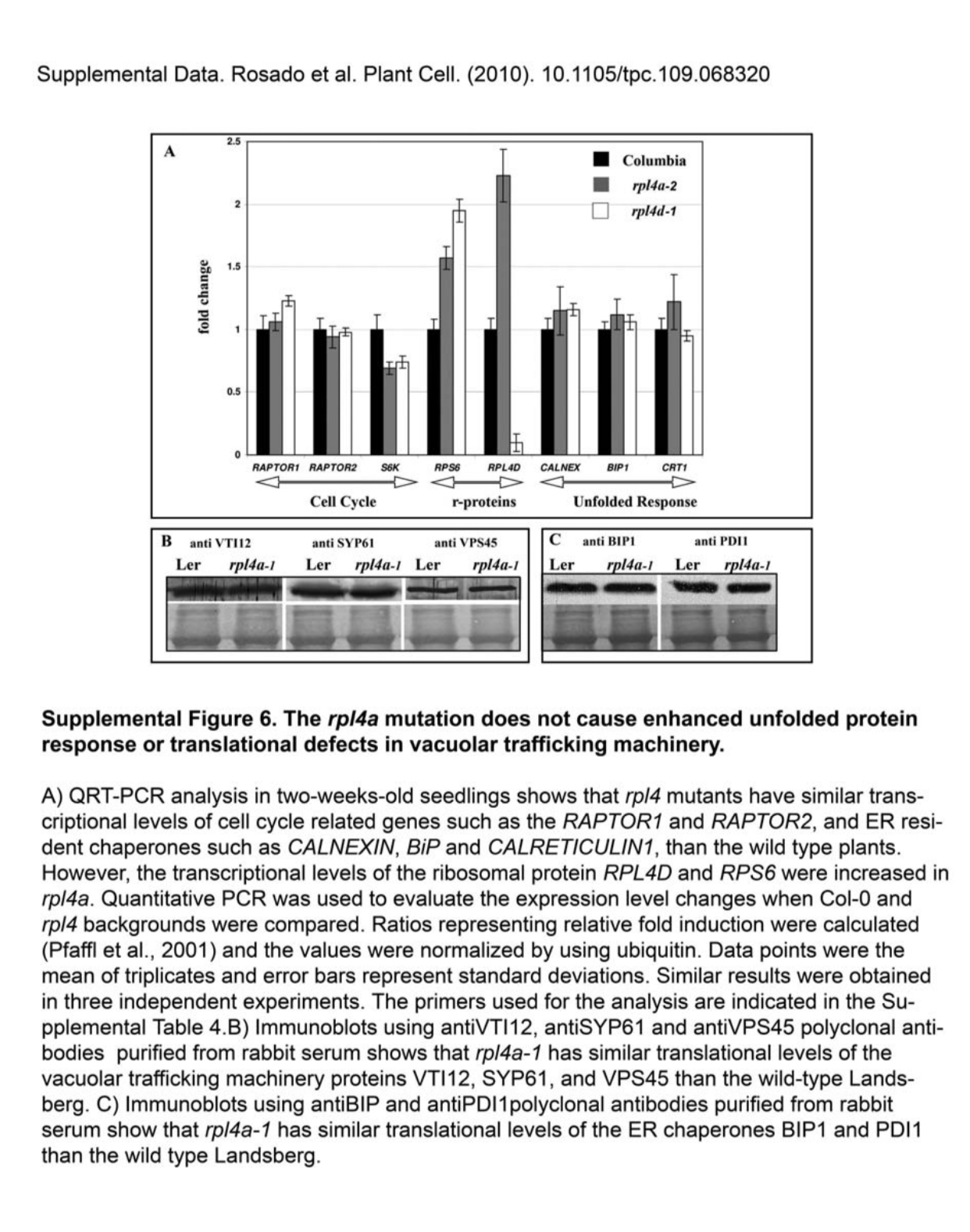

The Correction notice from March 2021 informs:

“The authors have discovered that an error occurred in the assembly of Supplemental Figure 6 panels B and C where it appears that loading controls were reused from Supplemental Figure 1.
The primary data and materials relevant to this work are no longer available. A.R. retrieved independent digital scans for 8 immunoblots that were either included (3) or discarded (5) during the assembly of Supplemental Figure 6B and C; however these scans do not provide sufficient information to assign the corresponding loading controls with confidence. Nevertheless, we stand by the results presented in Supplemental Figure 6B and C […]
We would also like to state that the results presented in Supplemental Figures 6 panels B and C do not affect, change, or interfere with any of the other findings and conclusions presented in the manuscript.“
The notice corrects one figure, but totally ignores the evidence in another. First author Abel Rosado is now associate professor at University of British Columbia in Vancouver, Canada. He hinted that the raw data for that figure was also not available, but provided a replacement experiment on PubPeer, from “a couple of additional repetitions that were used while optimizing the Figure 4A inmmunoblots.” It is truly mysterious why the authors diligently stored and catalogued the raw data for their unpublished “repetitions”, but not the raw data for the published figures which somehow disappeared.
Maybe this statement by Raikhel in an email [to LS] sheds some light:
“I am retired as of 2016 from UC Riverside and different PIs are working in my former lab now. Thus, not much documents are available there, however, people who did the work have some information and they are looking into accusations.“
Seems someone was very selectively trashing raw data at UC Riverside? Raikhel did not reply to an inquiry about that strange correction. Good to remember what she said before:
“neither I nor any of my colleagues are aware or would tolerate any data manipulation“
And now even The Plant Cell looked away from that wretched Figure 4, despite past experiences with their own top editors (here and here).
Did Plant Cell learn from Voinnet Affair?
The Plant Cell is an elite journal, its authors and editors are some serious heavyweights whose labs cannot be associated with data manipulation.
In Drakakaki et al. Cell Res 2012, Figure 1A carries an obvious lane duplication that was also horizontally flipped. The immunoblot analyses the sucrose gradient fraction enriched with the fluorescently tagged SNARE SYP61-CFP. Another Westen blot control in Figure 1, testing the purity of membranes, looks fishy too, and in Figure 1B no control for the immuno-EM was included. Figure 1 is crucial for proving the quality of the isolated membranes later used for the proteomic analysis. First author is Georgia Drakakaki, now associate professor at UC Davis.
Georgia Drakakaki , Wilhelmina Van De Ven, Songqin Pan , Yansong Miao, Junqi Wang , Nana F Keinath, Brent Weatherly , Liwen Jiang, Karin Schumacher, Glenn Hicks, Natasha Raikhel Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis Cell Research (2012) doi: 10.1038/cr.2011.129


Incidentally, a co-author of that paper, Karin Schumacher, Vice-rector for Quality Development at the University of Heidelberg, retracted 2 years ago the paper Patri-Nebioglu et al eLife 2019 soon after it was published, because of “irregularities associated with the western blots […] performed by Gorkem Patir-Nebioglu“. Initially, Schumacher and her first author and PhD student rebutted the concerns of data manipulation by posting on PubPeer what they declared to be raw data. It proved to be fabricated also.
Schumacher then admitted on PubPeer that “the blots in Figure 3E have indeed been manipulated in an inappropriate manner” but insisted: “the manipulation does not affect the outcome of the experiment.” She announced to replace the fake data with repeat experiments, having learned “a very painful lesson“.
The journal eLife announced an investigation, and days later, Schumacher announced a retraction and was saluted for her exemplary attitude. In any case, it was not the only falsified figure in that paper: also Figure 5B proved fishy.
Other papers co-authored by Schumacher still wait for their corrections, e.g. Pellisier et al Plant Phys 2004, also criticised on PubPeer:

But we return to the publications from Dr Raikhel’s lab. More panel duplications in Norambuena et al. BMC Chem Biol 2008, no SNAREs involved this time but dot-blot secretion assays, in an article consisting of three very simple main figures and two tables. So difficult not to mess it up with such an elaborate layout! First author Lorena Norambuena is now professor at University of Chile.
Lorena Norambuena, Jan Zouhar, Glenn R Hicks, Natasha V Raikhel Identification of cellular pathways affected by Sortin2, a synthetic compound that affects protein targeting to the vacuole in Saccharomyces cerevisiae BMC Chemical Biology (2008) doi: 10.1186/1472-6769-8-1
“Figure 2, treatment with different compounds (coded to the left) on S. cerevisiae to test the ability of each to trigger secretion of CPY. Dot-blot secretion assay using monoclonal antibodies raised against CPY. Some panels appear to be much more similar than expected (boxes of same color).“

Maybe Dr Norambuena’s colleague at that same university, Claudio Hetz, can advice her on the high art of figure assembly?
The Claudio Hetz Blues
“…Dr. Hetz seems rather to regret that he did not have better tools for editing the figures, so that the undeclared interventions would have gone unnoticed.” – University of Chile investigative report.
Another vintage paper, Ahmed et al. J Cell Biol 2000 , again a panel duplication:
Sharif U. Ahmed, Enrique Rojo, Valentina Kovaleva, Sridhar Venkataraman , James E. Dombrowski, Ken Matsuoka, Natasha V. Raikhel The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana The Journal of Cell Biology (2000) doi: 10.1083/jcb.149.7.1335

And from the same golden age, here is Sarria et al. Plant Physiol 2001:
Rodrigo Sarria , Tanya A. Wagner, Malcolm A. O’Neill, Ahmed Faik, Curtis G. Wilkerson , Kenneth Keegstra , Natasha V. Raikhel Characterization of a Family of Arabidopsis Genes Related to Xyloglucan Fucosyltransferase1 PLANT PHYSIOLOGY (2001) doi: 10.1104/pp.010596

It’s very shameful that nobody, including the first author Rodrigo Sarria (now R&D head at AgBiome), found time to correct this paper. Especially since Raikhel has back then just started as Editor-in-Chief of that journal Plant Physiology when the study was published.
Another putative panel duplication & flip in Zheng et al. Plant Physiol 2002, again published when Raikhel was Editor-in-Chief of that journal:
Haiyan Zheng , Sebastian Y. Bednarek , Anton A. Sanderfoot, Jose Alonso, Joseph R. Ecker, Natasha V. Raikhel NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE PLANT PHYSIOLOGY (2002) doi: 10.1104/pp.003970


No corrections were issued for these papers, despite Raikhel’s announcement form January 2021 [in an email to LS]:
“If we find any mistake we will definitely send a correction to a journal or deal with the issues accordingly.
But an Erratum was published in February 2021 for Rojas-Pierce et al. Chem Biol 2007 :
“In the originally published version of this article, we mistakenly duplicated lane 1 into lane 2 in Figure 5A. The revised Figure 5A appears below with the corrected lanes. This error does not affect the conclusions of this study, and the authors regret this error.”
Marcela Rojas-Pierce, Boosaree Titapiwatanakun, Eun Ju Sohn, Fang Fang, Cynthia K Larive, Joshua Blakeslee, Yan Cheng , Sean R Cutler, Wendy A Peer, Angus S Murphy, Natasha V Raikhel Arabidopsis P-glycoprotein19 participates in the inhibition of gravitropism by gravacin Chemistry & Biology (2007) doi: 10.1016/j.chembiol.2007.10.014
“Figure 5A, there is a band duplication (red boxes).“
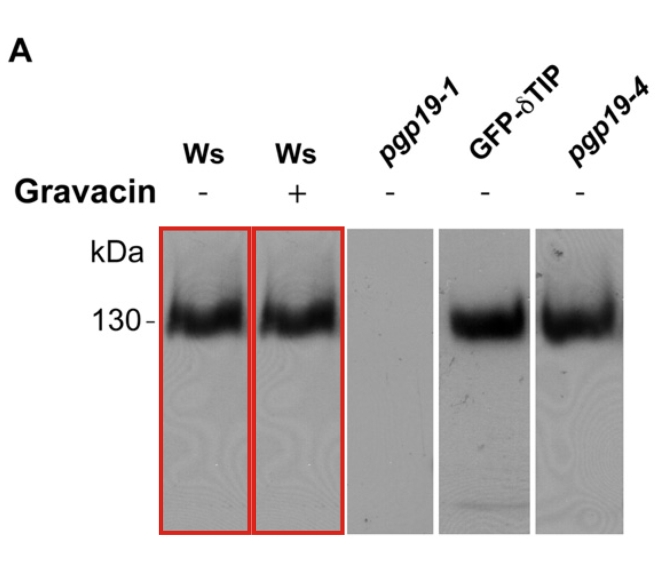
The first author Marcela Rojas-Pierce, now professor at the North Carolina State University, explained on PubPeer:
“there was an erroneous lane duplication in this figure, but I can assure you that this was an honest mistake. This particular blot was done in the laboratory of our collaborator, Dr. Angus Murphy, and files with scans were exchanged at the time via Fedex (as CDs). We tracked the original films from 2007 and concluded that the mistake arose when combining data from different blots and while removing one lane.”
Rojas-Pierce also wrote in an email [to LS]:
“I want to assure you that there was no intentional data manipulation of any kind. This was a combination of an honest mistake while preparing the figure and oversight from my part during paper submission.”
Indeed, that paper was corrected. Unlike the others.
In this regard, we end with the collaborative work Zhang et al. Mol Cell 2011 , with a blank panel duplicated five times. Its corresponding author is Hailing Jin, professor at UC Riverside, the institution where Professor Raikhel spent the last fifteen years or her long and productive career.
Xiaoming Zhang, Hongwei Zhao, Shang Gao, Wei-Chi Wang, Surekha Katiyar-Agarwal, Hsien-Da Huang, Natasha Raikhel, Hailing Jin Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(∗)-mediated silencing of a Golgi-localized SNARE gene, MEMB12 Molecular Cell (2011) doi: 10.1016/j.molcel.2011.04.010

Jin explained on PubPeer in January 2021:
“I was so shocked to see the duplication because I have been always very careful to double check all the original results and blots for publication. I am so sorry that I was not careful enough at checking the blank/negative blots this time, and I did not spot the big mistake of duplicated blank panels. After I saw your message, I immediately contacted the two First Authors, Xiaoming Zhang and Hongwei Zhao, to find the original blot images and to figure out how this mistake could have occurred. Luckily, Hongwei Zhao, my former postdoc who did all the AGO-IP experiments related to this figure, admitted that he made this terrible mistake, and managed to find the original U6 Northern blots (two biological replicates) and the original Western blot for the Tubulin control. […]
Hongwei Zhao explained to me why he made such a big mistake. He said during that time, he was extremely stressed out by his daughter’s upcoming heart surgery. He was distracted with all the doctor’s examinations and appointments for his daughter’s medical condition, and had to be off work regularly during that period of time. So he asked Xiaoming to help him develop the x-ray films and scan the phosphoimages for these experiments. He said when Xiaoming told him over the phone that these controls were blank with no bands, he used the image of AGO1 input blank to temporarily reserve the space for these controls and planned to replace them with the real images after he returned back to the lab. However, he forgot to replace them or tell Xiaoming about it after the operation, otherwise Xiaoming would have helped him finish the figure making. Hongwei feels very sorry about this, and as a PI now himself, he totally understands how much damage such careless mistakes can cause. It is a good lesson for all of us.“
9 months have passed since, and the paper still wasn’t corrected, maybe it suffices that the guilty parties have been named and made to apologise. Or maybe also because it’s Molecular Cell.
Fousteri affair: Dutch integrity thwarted by academic indecency
Two and a half years after Maria Fousteri was found guilty of scientific misconduct by her former employer, the Leiden University Medical Center (LUMC), exactly nothing at all happened. ERC and Molecular Cell ignored LUMC letters from June 2016, while Fouster’s British co-authors interfered to save own papers. Of 4 scheduled retractions, none took place.
A good lesson for all of us indeed.

Get For Better Science delivered to your inbox.
Make a one-time donation
Make a monthly donation
Choose an amount
Or enter a custom amount
Your contribution is appreciated.
Your contribution is appreciated.
DonateDonate monthly






An interesting question, perhaps worthy of scientific investigation: are people without any sense of shame have a tropism towards laboratory work, or does something in laboratories tend to erode that sense in (some of) those who work there?
Also: why are there so few scapegoats these days? Are they dying out for some reason?
LikeLike
“…or does something in laboratories tend to erode that sense in (some of) those who work there…”
This.
In science, I think the blank slate hypothesis applies most of the time: people inherently good that are corrupted by the environment. Here, it may be a difficult advisor, or the desire of a decent job, or to get a grant. Think Lord of the Flies.
Offer well paid, steady positions for post-docs, at the level of faculty, and the problem would go away.
LikeLike
Pingback: The curative plant extracts from Allahabad – For Better Science
An update on the dreadful paper Drakakaki et al. Cell Res 2012, which not only contains fabricated data, it is also lacking the most elementary controls, and doesn’t make any sense at all.
The Scientific & Managing Editor of the journal Cell Research, Dr. Lei Cheng, replied to my latest request of update.
Here her answer, received today March 2, 2022:
“Dear Dr …,
Since this paper has been published near 10 years and the first author, Georgia Drakakaki, has moved places, the corresponding author has long retired and the lab is closed. Thus, Dr Drakakaki told me that he/she will look into it and get back to me with the best he/she can. I will contact you once I received his/her reply. Thanks for your patience. Best, …”
This doesn’t sound very promising.
The journal was informed on January 2, 2021.
On July 26, 2021, Dr. Cheng replied to a previous request of update as follows:
“Dear Dr. …
It’s a pity that we did not receive the feedback from the corresponding author (–> Prof Raikhel). We checked the email address which we used is correct. We will try our best to get in touch with her and hope she can give us some explanation. Best”
I then provided Dr. Cheng the email address of first author Dr Drakakaki.
I also provided Dr. Cheng the email address of corresponding author Prof. Raikhel (Prof Raikhel used this address to communicate to Leonid some months ago), in case they had used a wrong/old one.
Obviously Prof. Raikhel is no longer interested to shed light to any problematic paper of hers. She didn’t reply ANYTHING to the journal, despite she promised to clarify the issues.
Now the editorial office is waiting for Dr. Drakakaki (professor at UC Davis !!!) to search (maybe listlessly?) for the data. The article was actually 8 years and 4 months old when I reported it to the journal.
Expect highest level of efficiency by Dr. Drakakaki, dear readers.
LikeLike
Pingback: Rick Vierstra probably meant well – For Better Science
9th paper flagged on PubPeer for Karin Schumacher:
https://pubpeer.com/publications/4F1B9DF4F7A0270E3C1ECF3C0EBDEC
After the 2019 retraction of Patir-Nebioglu_et_al Elife paper (whose issues she initially denied on PubPeer), she was rewarded with the position of Vice-rector for Quality Development (!) at Uni Heidelberg. Theoretically her office would terminate in a few weeks, but she could get an extension of it. We’ll see.
LikeLike
Terrible erratum to Sanderfoot et al. 2001, dated 1 Octoer 2021: https://www.molbiolcell.org/doi/10.1091/mbc.12.12.3733-corr?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed
LikeLike
Yet another deadful correction, this time to Drakakaki et al. 2012 Cell Res, dated 21 September 2023: https://www.nature.com/articles/s41422-023-00865-5
“After concerns were raised by readers regarding some SYP51-CFP immunoblot features in Fig. 1A, the authors were unable to retrieve the original image after 13 years of publication for validation; therefore, an alternative image is provided in the revised Fig. 1. The authors, in addition, have provided in Fig. 1B (iii, iv) control images for immuno-negative staining that were not included in the original publication.”
I cannot help but notice that in corrected Figure 1A the upper blot was replaced, whilst the middle and lower blots remained the same but also replaced with versions of better resolution. Does that mean that for two blots out of three the authors were able to retrieve the original scans, but not for the upper one where the lane duplication was spotted? Moreover, where does the replacement come from? Did the authors performed a completely new experiment, or did they retrieve an second try from the past?
Of note, this group is used to issue strange corrections to problematic data: https://pubpeer.com/publications/ED775781540737C87DBABC987C54AE https://pubpeer.com/publications/AD38B076BEB859957C5F84511A3B5E
LikeLike
https://www.plantsciences.ucdavis.edu/people/georgia-drakakaki
doing well in US academia. Not a failed scientist.
LikeLike