Clare Francis recently investigated another set of French scientists, and posted it on PubPeer. One of them is the cancer researcher Anne Dejean- Assemat, Research Director Classe Exceptionnelle at INSERM and professor at the Institut Pasteur in Paris. The other is her German mentee Oliver Bischof, now group leader under Dejean’s leadership. It is very likely Bischof is being groomed to succeed Dejean when she retires. And to be fair, the way this Pasteur lab is run, he is definitely the right man for the job.
This is Dejean’s CV, as presented by her “Nuclear Organization and Oncogenesis” lab at Institut Pasteur:
“Anne Dejean is Research Director at INSERM, Professor at the Institut Pasteur and Head of the Laboratory of Nuclear Organization and Oncogenesis/INSERM U993. She graduated from Pierre et Marie Curie University in Paris and earned her PhD in Pierre Tiollais’ lab at the Institut Pasteur in 1983. Member of EMBO and of the French Academy of Sciences, she has received the Gagna and Van Heck Prize in 2003, the L’Oréal-UNESCO for Women in Science Awards in 2010, the Grand Prix INSERM in 2014 and the Sjöberg Prize in 2018. She was awarded two ERC Advanced Grants, in 2011 and 2018.”
I would like to show you how exactly Professor Dejean achieved so much. And yes, I know European Research Council (ERC) leadership doesn’t believe in research integrity, but still. The scientific community has a right to know who and how receives such exclusive public research funding, which honest scientists see routinely denied. Dejean’s new €2.5mn Advanced Grant “Deconstructing the role of SUMO on chromatin in cell identity and tissue repair” was awarded in 2018, and don’t worry, it will continue for the next 5 years excellently. On this, you can trust ERC. Carlos Lopez-Otin enjoys his ongoing grant despite retractions, mouse murder, admitted data fakery and cessation of research activities, and even proven the fraudster Maria Fousteri was free to use up all of €2mn to her pleasure, after the university notified the ERC.

Now, someone, or a group of someones in Dejean’s lab has been naughty. We cannot know who, but the evidence of their naughtiness is there. Let’s look at this almost 20 year old paper:
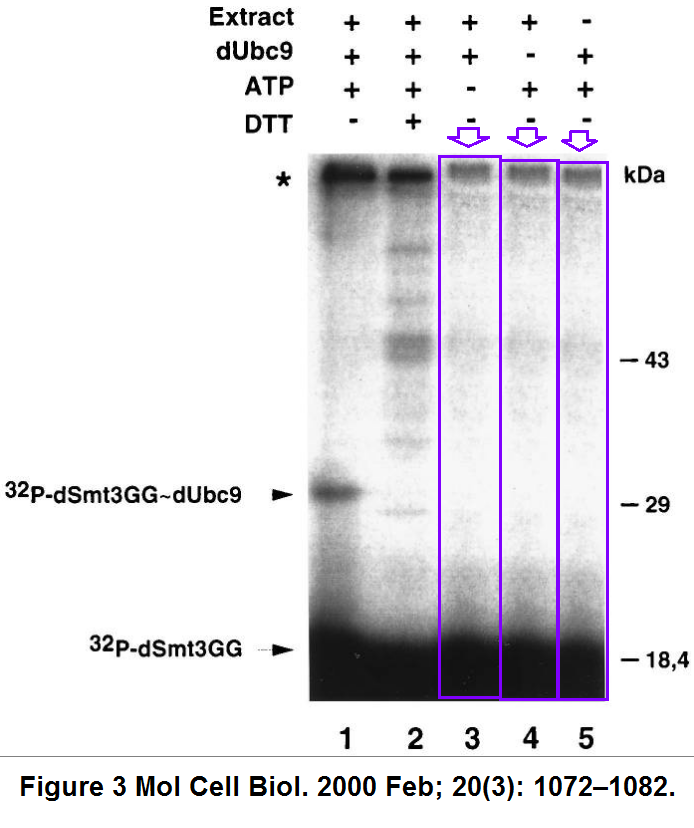
F. Lehembre, P. Badenhorst, S. Müller, A. Travers, F. Schweisguth, A. Dejean Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies Molecular and Cellular Biology (2000) doi: 10.1128/mcb.20.3.1072-1082.2000
Normally, gel lanes do not triplicate like this, unless the authors disagreed with their actual experimental results and decided prove them wrong, using The Photoshop, or in this case, probably the good old scissors and glue. Maybe the issue is serious enough for the journal to overrule the 6-year deadline strategy? After all, the journal MCB already had to correct a Dejean-coauthored paper from INSERM in Paris, for duplicated data (Erker et al MCB 2013).
Or how does this happen, in the same journal, but 5 years later? Something horrible happened to Figure 3C in 2005, this time it was most likely the revolutionary biomedical research tool of Photoshop being applied:
O Bischof, K Nacerddine, A Dejean Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways Molecular and Cellular Biology (2005) doi: 10.1128/mcb.25.3.1013-1024.2005
How come 3 out of 4 bands of the top PML western blot look so similar, especially after a rotation? Also the two central E7 bands seem to be mirror images of each other, and it turned out, also the first two p53 bands are strangely similar, and the same applies for the CBP bands below.
How exactly did Dejean and Bischof plan to cure leukaemia with this technology? One year later, they published another paper with similar issues, where some of Bischof et al MCB 2005 data was apparently reused in excitingly new context. Luckily the new journal was Molecular Cell, this Elsevier outlet seems to have an unhealthy obsession of attracting and cultivating manipulated data.
O Bischof, K Schwamborn, N Martin, A Werner, C Sustmann, R Grosschedl, A Dejean The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis Molecular Cell (2006) doi: 10.1016/j.molcel.2006.05.016

Now, what do we have here? A set of 3 bands from Figure 2C of MCB 2005 which used to be p53, but then somehow became Rb, a very different protein. One of this trio of jumping bands grabbed another friend and cloned themselves again to become yet another protein, Cyclin D1 in Figure 4E of Molecular Cell. It is not helpful that the sample legend is different for every single instance. And since we are discussing the magical permutation of p53 into Rb protein, let’s look at Figure 4A:

The band even got hyper-phosphorylated while transforming itself from p53 to pppRb. There are more gems in that 2006 paper, have a look at Figure 6H:
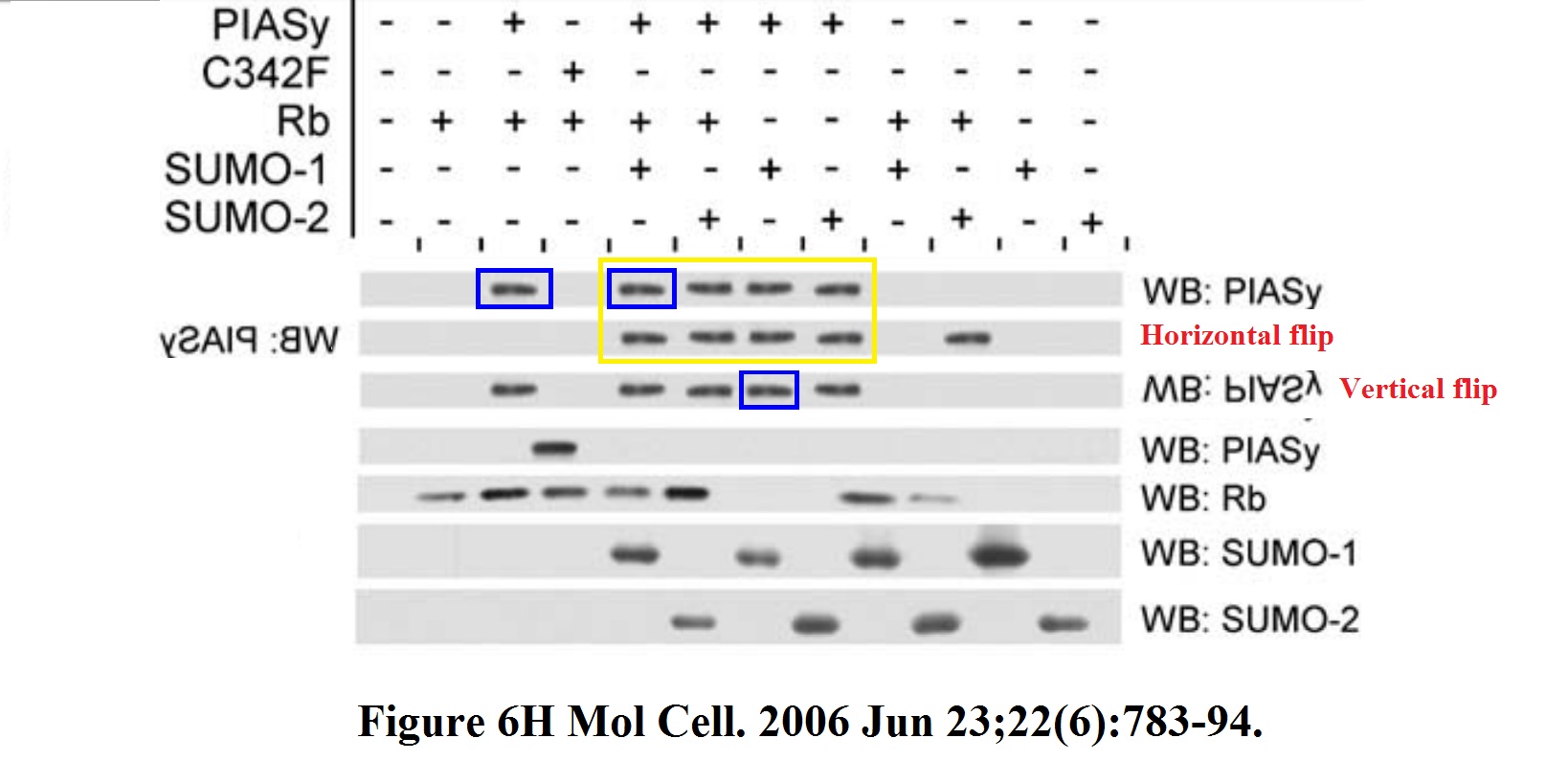
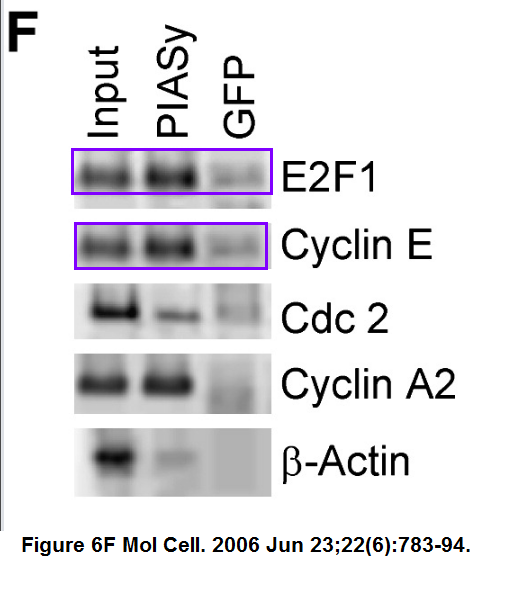
Can it be that top western blot panel shows the same band all over, copy-pasted with an occasional flip? Was someone taking the proverbial PIAS here? Also the Figure 6F has what Institut Pasteur experts will probably call “an inadvertent duplication”.
Now, Clare Francis did report all these and other issues to Molecular Cell editors, but it is rather like writing to Philip Morris to complain about someone smoking in a restaurant. This Cell Press journal chose to do nothing at all about fraudulent papers by former CNRS president Anne Peyroche, and rejected retraction requests in the case of Maria Fousteri.
Before the first author of this Photoshoppian masterpiece joined the Dejean lab in Paris, the German Oliver Bischof did a brief postdoc stint at the lab of Farzin Farzaneh at King’s College London. There was much to learn also: Farzaneh was recently investigated for data manipulation, but no research misconduct was found, only “poor research practices” which required Errata in 5 journals.
Between Farzaneh and Dejean postdoc, Bischof did a stint at the Berkeley lab of Judith Campisi, a star researcher in the field of cellular senescence. This collaboration moved the research area ahead with this important contribution:

O Bischof, S Galande, F Farzaneh, T Kohwi-Shigematsu, J Campisi Selective cleavage of BLM, the bloom syndrome protein, during apoptotic cell death The Journal of biological chemistry (2001) doi: 10.1074/jbc.m006462200
Now this cannot have happened by incident, no matter how drunk The Bischof might have been, because the upper and lower set of bands are supposed to have been on one solid gel. No way the upper 20/46 bands can accidentally become lower 20 set, flipped.
The shenanigans apparently continued in Paris. This last-author paper from Institut Pasteur Bischof proudly shows among his selected publications:
M Ogrunc, RI Martinez-Zamudio, P Ben Sadoun, G Dore, H Schwerer, P Pasero, JM Lemaitre, Anne Dejean, O Bischof USP1 Regulates Cellular Senescence by Controlling Genomic Integrity Cell Reports (2016) doi: 10.1016/j.celrep.2016.04.033
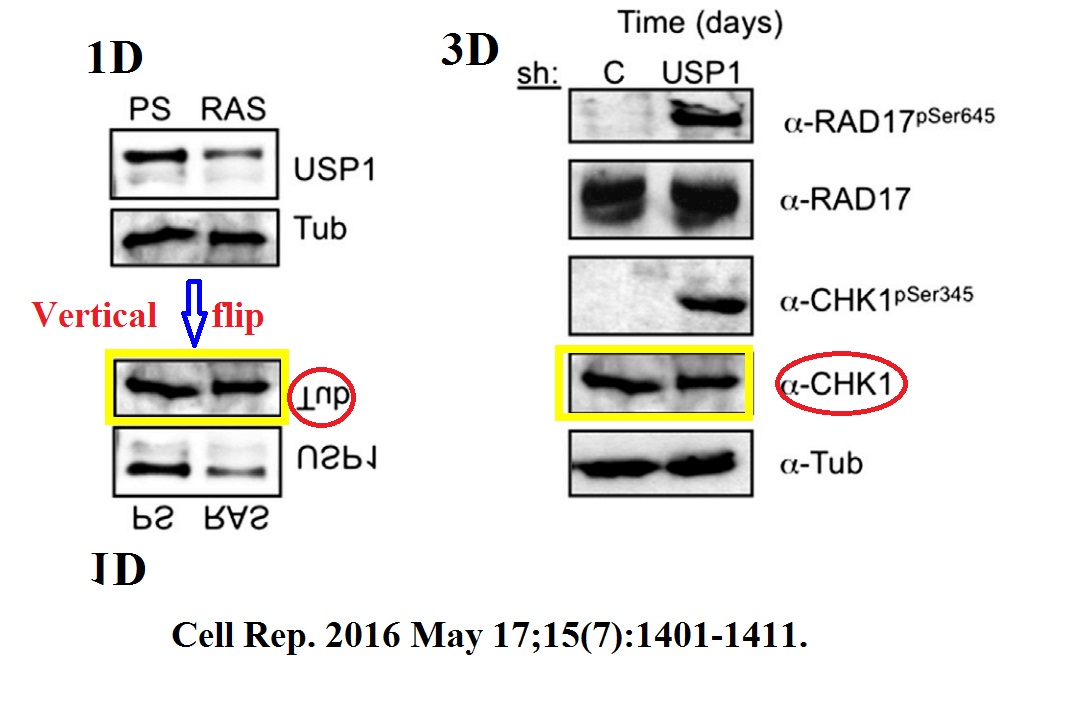
Now I should immediately declare that the first author Müge Ogrunc and I worked for several years together at the same DNA damage research small lab at IFOM in Milan, Italy. I tried to contact Müge about this copy-pasted and flipped CHK-1 western blot, but received so far no feedback. But then again, my former colleague Müge joined the Dejean-Bischof lab at Pasteur only for two years, 2015-2017, while the data integrity issues go way back there.
Dejean is now 62 and will probably retire in a couple of years, maybe when her new ERC grant ends. With Bischof, she prepared a worthy successor to take over her artisan craft business, which now started to conquer new sources of grant cash: ageing or senescence research. The general questionable research ethics in Dejean lab shine through in examples like this:
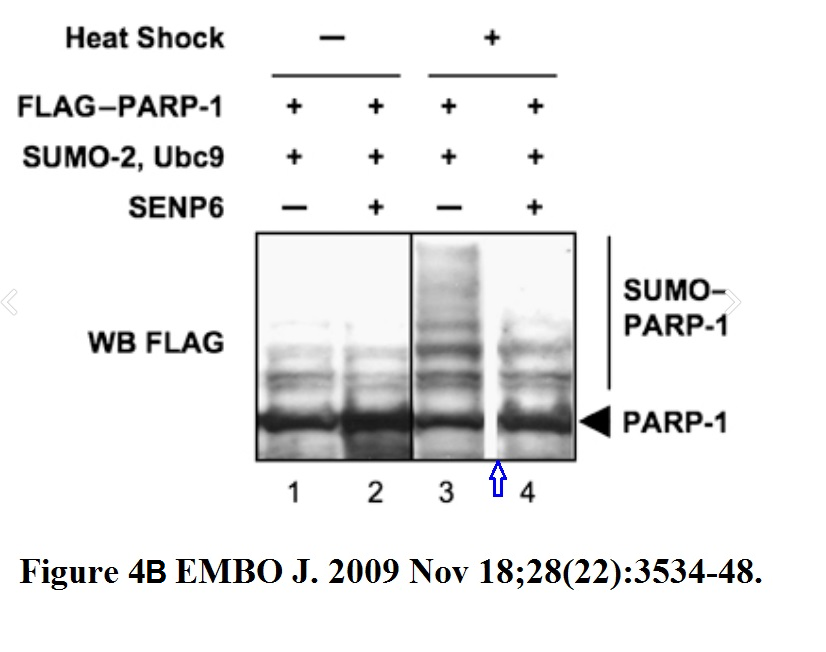
N Martin, K Schwamborn, V Schreiber, A Werner, C Guillier, XD Zhang, O Bischof, J Seeler, A Dejean PARP-1 transcriptional activity is regulated by sumoylation upon heat shock The EMBO Journal (2009) doi: 10.1038/emboj.2009.279
Now the paper is 10 years old, and the French and German authors might declare that they missed previous guidelines in biomedical publishing where gel splicing should be avoided, and where not possible, clearly indicated, because the guidelines were in English. But thing is, in Figure 4B they did provide a clear black vertical line to indicate splicing in one case, but they chose not to do this in other cases. Why? Because you cannot compare the signal of a + vs – treatment on a spliced gel. In brief, the authors bullshitted the EMBO editor, the peer reviewers and the scientific community while knowing perfectly well that what they did was not right.
Or how about another paper from same time, with same key authors:
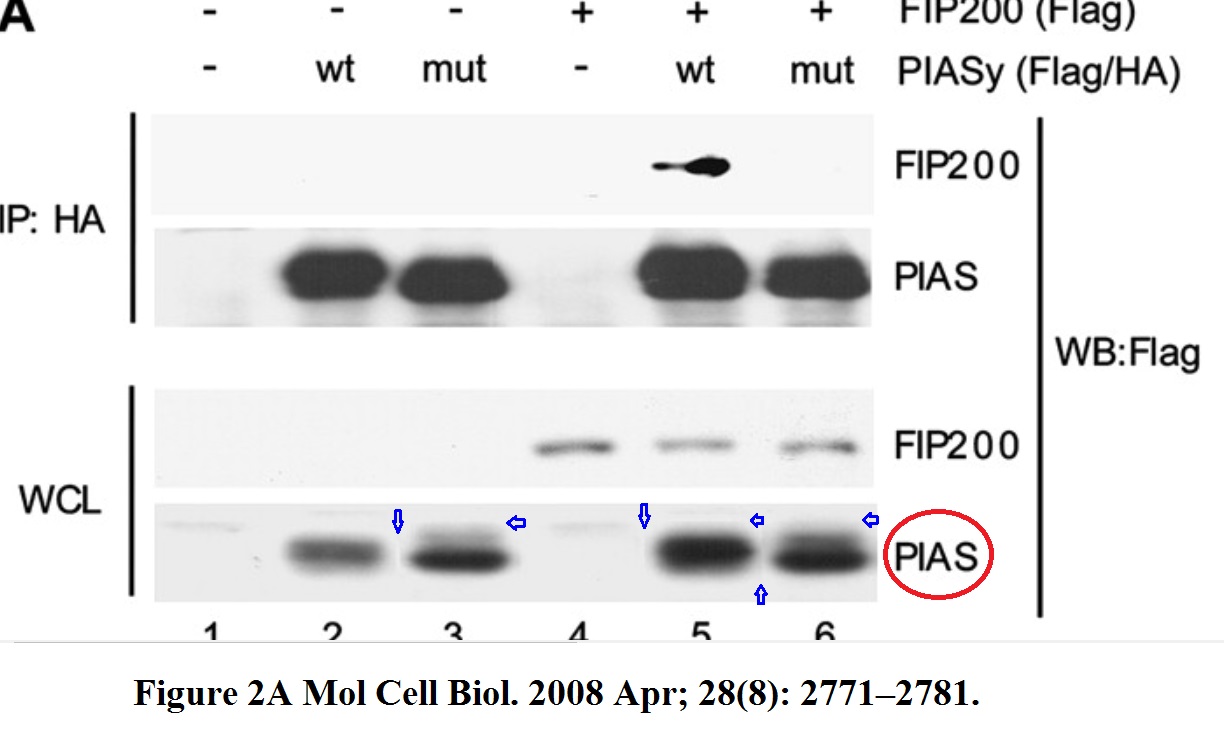
N Martin, K Schwamborn, H Urlaub, B Gan, JL Guan, A Dejean Spatial interplay between PIASy and FIP200 in the regulation of signal transduction and transcriptional activity Molecular and Cellular Biology (2008) doi: 10.1128/mcb.01210-07
Now it might appear like some sloppy but otherwise innocent gel splicing, but it is not. The horizontal splice edges are the giveaway that something more sinister than mere excision of irrelevant gel lanes took place. The PIAS panel in Figure 2A is apparently a collage of various gel bands slapped on in Photoshop, the same thing happened to the PIAS panel of Figure 3C. There however, the bands were not just stuck on randomly: they were precisely arranged in Photoshop on a ladder, to indicate the transgenic vs endogenous PIAS protein. Basically, another PIAS-taking exercise by Anne Dejean and her lab, and yet another reason for the publisher American Society for Microbiology (ASM) to investigate these older works in their journal MCB.
The following is a collaborative paper from the Institut Pasteur, maybe Dejean can check if the problematic figures were made in her lab.
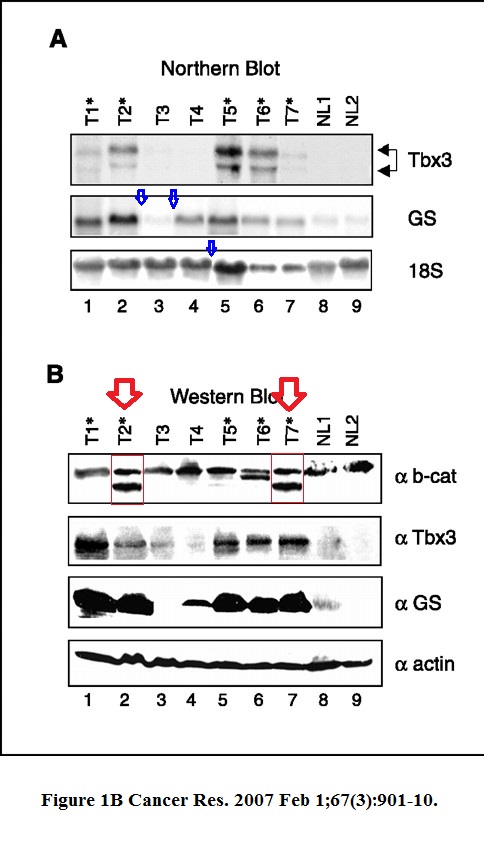
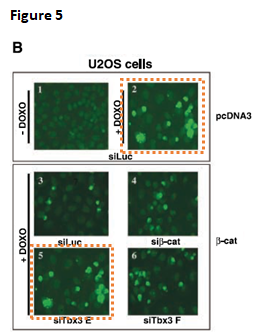
CA Renard, C Labalette, C Armengol, D Cougot, Y Wei, S Cairo, P Pineau, C Neuveut, A De Reyniès, A Dejean, C Perret, MA Buendia Tbx3 is a downstream target of the Wnt/beta-catenin pathway and a critical mediator of beta-catenin survival functions in liver cancer Cancer Research (2007) doi: 10.1158/0008-5472.can-06-2344
Not just some splicing, a b-cat band was most obviously cloned in Figure 1B. Even Nature, where Dejean published quite a lot, was not safe. That journal however replied to Clare Francis and announced to look into the case. On its own, there is just a duplicated SUMO gel:
D Ribet, M Hamon, E Gouin, MA Nahori, F Impens, H Neyret-Kahn, K Gevaert, J Vandekerckhove, A Dejean, P Cossart Listeria monocytogenes impairs SUMOylation for efficient infection Nature (2010) doi: 10.1038/nature08963

But considering what else went on and still goes on in the Dejean and now Dejean-Bischof lab? Well, the worst things ended up strategically in Molecular Cell. We are dealing with real professionals here.
INSERM is already well aware of the PubPeer evidence. Clare Francis also informed Institut Pasteur, and its president Stewart Cole replied to him:
“As is our practice in such matters I have instructed the Committee for Scientific Integrity to conduct an enquiry into the allegations. This will take some time. The PIs concerned have been informed.”
Now I would not get overexcited here. Institut Pasteur previously investigated another affair, relating to their collaboration with the present rector of Karolinska Institutet, and found no data manipulation despite authors admitting a possible gel lane triplication, while the alleged original data did not match.
My prediction: conclusions not affected, original data unfortunately eaten by some goats.
Update 25.05.2021
Bischof has 4 retractions now, 3 with Dejean (Bischof et al MCB 2005, Bischof et al Mol Cell 2006, Ogrunc et al Cell Reports 2016) and one from his postdoc period with Judith Campisi (Bischof et al JBC 2001). Today, I received this email from Christophe d’Enfert, scientific director of Institut Pasteur:
“An enquiry into the alleged case of scientific misconduct by Dr. Oliver Bischof, a CNRS scientist, and Prof. Anne Dejean was launched in the fall of 2019 and recently concluded. Prof. Anne Dejean’s unit at Institut Pasteur is affiliated with Inserm. This enquiry was thus conducted jointly by the scientific integrity officers of the CNRS, Inserm and the Institut Pasteur. Several of the publications linked to the allegation were retracted prior to completion of the enquiry (PMID: 33626350, PMID: 33338405, PMID: 33277404, PMID: 33106371).
The team in charge of the enquiry presented a written report to the heads of the three organizations concerned. The evidence and conclusions indicate that during the preparation of many of the publications O. Bischof, but not A. Dejean, had engaged in fraudulent practices, in violation of the rules of honesty and scientific integrity. After due consultation with the President of the CNRS, Prof Cole, President of the Institut Pasteur, asked O. Bischof to leave the Institut Pasteur without delay. This is why the group of Oliver Bischof is not listed anymore.“

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism, however small it appears to you, will greatly help me with my legal costs.
€5.00



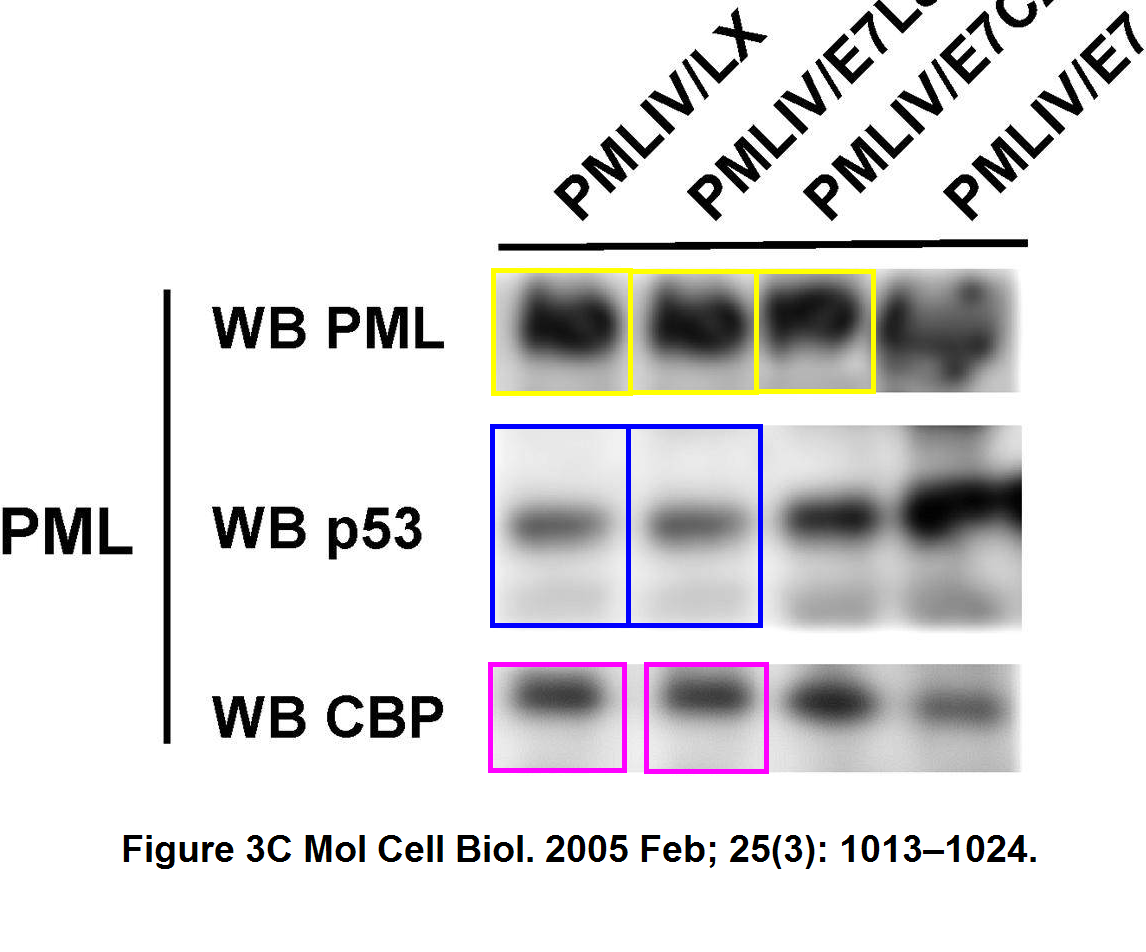



“How come 3 out of 4 bands of the top PML western blot look so similar, especially after a rotation?”
They look like little bunnies to me and they would be improved by googly eyes.

LikeLike
“Frédérique Vidal
@VidalFrederique
[Communiqué] Félicitations à Anne Dejean-Assémat et Hugues de Thé, lauréats du prix #Sjöberg 2018 décerné par l’Académie royale suédoise pour leurs recherches sur la leucémie aiguë promyélocytaire, une des formes les plus agressives du cancer du sang”
https://www.gouvernement.fr/ministre/frederique-vidal
http://retractiondatabase.org/RetractionSearch.aspx?AspxAutoDetectCookieSupport=1#?AspxAutoDetectCookieSupport%3d1%26auth%3dVidal%252c%2bFrederique
1 retraction (coauthor)
Reasons
+Concerns/Issues About Data
+Duplication of Image
+Manipulation of Images
1 Expression of Concern (first author).
Reasons
+Concerns/Issues About Image
+Investigation by Journal/Publisher
LikeLike
Nat Cell Biol. 2007 Jan;9(1):45-56. Epub 2006 Dec 17.
Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus.
Kumar PP1, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S.
Author information
https://www.nature.com/articles/ncb1516#author-information
Pavan Kumar P. and Oliver Bischof: These authors contributed equally to this work.
Affiliations
National Centre for Cell Science, Ganeshkhind, Pune, 411007, India
Pavan Kumar P., Prabhat Kumar Purbey, Dimple Notani & Sanjeev Galande
Unité d’Organisation Nucléaire et Oncogénèse/INSERM U579, Institut Pasteur, 28, rue du Docteur Roux, Paris, 75724, Cedex 15, France
Pavan Kumar P., Oliver Bischof & Anne Dejean
Max-Planck-Institute for Biophysical Chemistry, Bioanalytical Mass Spectrometry Group, Am Fassberg 11, Goettingen, D-37077, Germany
Henning Urlaub
Corresponding author
Correspondence to Sanjeev Galande.
https://pubpeer.com/publications/687186C326B8E8EB55BC33CE08DB35
Figure 5a.
LikeLike
Nat Cell Biol. 2007 Jan;9(1):45-56. Epub 2006 Dec 17.
Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus.
Kumar PP1, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S.
made me curious and I did some “lineage tracing”.
PLoS Biol. 2010 Jan 26;8(1):e1000296. doi: 10.1371/journal.pbio.1000296.
Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner.
Notani D1, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, Mehta S, Purbey PK, Joseph J, Galande S.
Author information
1
National Centre for Cell Science, Ganeshkhind, Pune, India.
Figure 1D. Much more similar than you would expect.
LikeLike
J Biol Chem. 2001 Apr 13;276(15):12068-75. Epub 2001 Jan 11.Selective cleavage of BLM, the bloom syndrome protein, during apoptotic cell death.Bischof O1, Galande S, Farzaneh F, Kohwi-Shigematsu T, Campisi J.Author information1Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley California 94720, USA.
Figure 5E. Much more similar after horizontal flip than you would expect.
LikeLike
Additional problematic data (image duplication) J Biol Chem. 2001 Apr 13;276(15):12068-75.
Figure 4B. Much more similar than you would expect.
LikeLike
Yet more problematic data J Biol Chem. 2001 Apr 13;276(15):12068-75.
Figure 5D.
See: https://pubpeer.com/publications/C63F1964D529BF3ECDFA1F52D15342#3
It takes time for things to come to light.
LikeLike
Exp Cell Res. 2000 Mar 15;255(2):135-43.
Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9.
Author links open overlay panelWeidongXuabcLiminGongdMaher M.Haddadabc Oliver Bischof e Judith Campisi e Edward T.H.YehdEstela E.Medranoabc1
a
Roy M. and Phyllis Gough Huffington Center on Aging, Baylor College of Medicine, One Baylor Plaza M320 and VAMC, Houston, Texas, 77030
b
Department of Cell Biology, Baylor College of Medicine, One Baylor Plaza M320 and VAMC, Houston, Texas, 77030
c
Department of Dermatology, Baylor College of Medicine, One Baylor Plaza M320 and VAMC, Houston, Texas, 77030
d
Institute of Molecular Medicine, University of Texas–Houston Health Science Center, Houston, Texas, 77030
e
Department of Cell and Molecular Biology, Lawrence Berkeley National Laboratory, University of California, Berkeley, California, 94720
Figures 4a and 5A. Much more similar than you would expect.
LikeLike
https://www.buckinstitute.org/lab/campisi-lab/
Judith is a big shot in the aging field. Note the slick front page of her lab website, and her powerful look of self-importance sigh
she is a journal editor with David Sinclair, who is notorious for selling his company, Sirtis to GSK for a lot of money in exchange for useless molecules (analogs of resveratrol) face palm
All we need is Aubrey DeGrey to round out this picture….
LikeLike
“All we need is Aubrey DeGrey to round out this picture….”
https://forbetterscience.com/2021/09/01/bleedem-while-theyre-young/
LikeLike
Is Judith Campisi a bit like Karen Vousden? Always there, all the time, “science lite”.
Famous for discovering one of the main markers (there are many markers) of cellular “aging”.
LikeLike
Not sure. I can say in her defense, Judith does write good review articles….
LikeLike
Judith Campisi is so powerful in the field of ageing/senescence you can’t imagine. How you dare you combing her papers!
She is up there with George Church.
LikeLike
“Oliver Bischof did a brief postdoc stint at the lab of Farzin Farzaneh at King’s College London.”
Farzin Farzaneh, King’s College London is penultimate author on both papers below, way to go!
Data in Toxicol Lett. 2009 Dec 15;191(2-3):118-22 looks very like data in J Leukoc Biol. 2005 Aug;78(2):503-14, yet experiments different.
Toxicol Lett. 2009 Dec 15;191(2-3):118-22. doi: 10.1016/j.toxlet.2009.08.012. Epub 2009 Aug 19.
RACK-1 overexpression protects against goniothalamin-induced cell death.
Inayat-Hussain SH1, Wong LT, Chan KM, Rajab NF, Din LB, Harun R, Kizilors A, Saxena N, Mourtada-Maarabouni M, Farzaneh F, Williams GT.
Author information
1
Toxicology and Biocompatibility Laboratory, Faculty of Allied Health Sciences, Universiti Kebangsaan Malaysia.
J Leukoc Biol. 2005 Aug;78(2):503-14. Epub 2005 May 3.
Functional expression cloning reveals a central role for the receptor for activated protein kinase C 1 (RACK1) in T cell apoptosis.
Mourtada-Maarabouni M1, Kirkham L, Farzaneh F, Williams GT.
Author information
1
School of Life Sciences, Keele University, Keele, ST5 5BG, UK.
LikeLike
Leukoc Biol. 2005 Aug;78(2):503-14 seems to have problems of its own,
Figure 7b. Very similar after re-sizing.
https://imgur.com/M1dVoX1
LikeLike
“O Bischof, K Schwamborn, N Martin, A Werner, C Sustmann, R Grosschedl, A Dejean The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis Molecular Cell (2006) doi: 10.1016/j.molcel.2006.05.016”
Figures 4A. Much more similar than you would expect.
LikeLike
Some more problematic data Mol Cell. 2006 Jun 23;22(6):783-94.
The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis.
Bischof O1, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A.
Author information
Nuclear Organisation and Oncogenesis Unit, INSERM U579, Institut Pasteur, 75724 Paris Cedex 15, France.
Figure 4E. Much more similar than you would expect.
LikeLike
“D Ribet, M Hamon, E Gouin, MA Nahori, F Impens, H Neyret-Kahn, K Gevaert, J Vandekerckhove, A Dejean, P Cossart Listeria monocytogenes impairs SUMOylation for efficient infection Nature (2010) doi: 10.1038/nature08963”
Correction announced by senior author at Pubpeer
https://pubpeer.com/publications/DA8AD8F94E6903DF242D6758D0C7B3#2
LikeLike
2020 correction published for Nature. 2010 Apr 22;464(7292):1192-5. doi: 10.1038/nature08963.
https://www.nature.com/articles/s41586-020-2154-7
Author Correction
Published: 16 April 2020
Author Correction: Listeria monocytogenes impairs SUMOylation for efficient infection
David Ribet, Mélanie Hamon, Edith Gouin, Marie-Anne Nahori, Francis Impens, Hélène Neyret-Kahn, Kris Gevaert, Joël Vandekerckhove, Anne Dejean & Pascale Cossart
In Fig. 4d of this Letter, the band corresponding to free SUMO1 in the left-hand blot is incorrect, as it is a repeat of the band corresponding to free SUMO2 in the right-hand blot. Figure 1 of this Amendment shows the original incorrect panel and the corrected panel of Fig. 4d, for transparency to readers. The full uncropped blots are shown in the Supplementary Information to this Amendment. The original Letter has not been corrected.
LikeLike
One of Dejean’s co-authors (Sanjeev Galande) is accumulating his own PubPeer list of questionable figures. https://en.wikipedia.org/wiki/Sanjeev_Galande
These papers are flagged now:
https://pubpeer.com/publications/336CC6E37960298F29D4FDA4188A6A (possible splicing)
https://pubpeer.com/publications/687186C326B8E8EB55BC33CE08DB35 (possible band re-use)
https://pubpeer.com/publications/D86B5754724C9235F612C04828167C (possible band re-use) “Ghost Shadow” paper
https://pubpeer.com/publications/A0686DD55CB3F80E743EFF7B8B1817 (possible splicing)
https://pubpeer.com/publications/C63F1964D529BF3ECDFA1F52D15342 (possible band manipulation/re-use)
https://pubpeer.com/publications/F1D6664B98B63D614922CA016D77D9 (possible splicing and band re-use)
https://pubpeer.com/publications/0BAC92DC47986918B4F41393EE893D (possible band manipulation/re-use)
Techniques learned at the feet of a master? Or are there more explanations similar to the “Ghost Shadow” excuse (below) that we can look forward to?
“These are ‘ghost’ shadows created by the thermal printer attached to the gel documentation system at that time. Any high intensity band has shown these white shadows on the side. This printer was also found to create many other artifacts and hence discontinued.” Sanjeev Galande 11/28/19
Note: Most of these were posted by other PubPeer anonymous peers. I contributed a few small items which I found independently. I haven’t made a systematic search through his papers. ~Cheshire
LikeLike
https://journals.asm.org/doi/10.1128/mcb.00040-22
Molecular and Cellular Biology
Correction
17 March 2022
Authors: P. Pavan Kumar, Prabhat Kumar Purbey, Dyavar S. Ravi, Debashis Mitra, Sanjeev Galande https://orcid.org/0000-0002-7251-1905 sanjeev@iiserpune.ac.in
Author correction.
“. Page 1624, Fig. 2C, lower panel: Lanes 4 and 5, representing the input of the ChIP reaction amplified using the IL-2-P2 primer pair, appear identical to the input control panel (lanes 1 and 2) using the IL-2-P1 primer pair for Fig. 7G. Both are supposed to represent equal amounts of chromatin, equivalent to loading controls.
Page 1628, Fig. 7G: The control panel (Mouse-IgG on top and Rabbit-IgG in the middle) for the ChIP-PCRs is inadvertently duplicated. Because of the time elapsed since publication, the original gel images are no longer available. This duplication does not change the overall conclusions of the article. We apologize for the confusion it might have caused.”
As Deiratonotus cristatus commented April 2022https://pubpeer.com/publications/D86B5754724C9235F612C04828167C#8
“MCB posted the correction with these statements. What is the correction here? Just a statement that the gel bands were inadvertently duplicated.”
LikeLike
What is going on with this senescence publication?https://pubpeer.com/publications/8DF6E66FEFAA2C3C7D5BD9C3FC45A2
LikeLike
https://www.sciencedirect.com/science/article/pii/S2352396419308102
LikeLike
Confused. Does it mean Kirkland’s patented senolytics work only when a Corrigendum of falsified data is rectally inserted?
https://www.technologyreview.com/s/612943/a-cell-killing-strategy-to-slow-aging-passed-its-first-test-this-year/
LikeLike
EMBO J. 2002 Jul 1;21(13):3358-69.
Deconstructing PML-induced premature senescence.
Bischof O1, Kirsh O, Pearson M, Itahana K, Pelicci PG, Dejean A.
Author information
1
Unité de Recombinaison et Expression Génétique, INSERM U 163, Institut Pasteur, 28 rue du Dr Roux, 75724 Paris Cedex 15, France.
Figure 6D.
See: https://pubpeer.com/publications/302B876419B626E90972A6CABA9AB7#2
LikeLike
In my understanding, the mentioned papers use western blotting. Proteins separated on gels are transfered (bloted) on a membrane. The membrane is then revealed using different “probes” made of labeled antibodies. As the SAME membrane is reused many times with different probes, this is no surprise that the specific shapes of bands is conserved between different figures!!!
LikeLiked by 1 person
Your IP address is in Paris. Is this why you try to troll here anonymously?
LikeLike
Sorry, I replied too quickly and it was a mistake. The western Blot rehybridation may explain some but not all images duplicates and flips. Moderators can delete my comment. Thanks
LikeLike
In different papers to represent different proteins/experiments..
LikeLike
Not a single reply on Pubpeer and now this. I am giving up.
LikeLike
Who would guess that his new NCB paper has data duplication?
https://pubpeer.com/publications/F4CAE86DB084A728CA076FDB56B5D6
LikeLike
Pingback: Münster charges Jens Schwamborn’s mentor Andreas Püschel with research misconduct, again – For Better Science
2020 retraction for:
Mol Cell Biol . 2005 Feb;25(3):1013-24. doi: 10.1128/MCB.25.3.1013-1024.2005.
Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways
Oliver Bischof 1, Karim Nacerddine, Anne Dejean
Affiliations collapse
Affiliation
1Nuclear Organisation and Oncogenesis Unit, INSERM U579, Institut Pasteur, 28 rue du Dr. Roux, 75724 Paris Cedex 15, France.
PMID: 15657429
2020 retraction notice.
https://mcb.asm.org/content/40/22/e00482-20
“We hereby retract this article. We were recently made aware of the presence of irregularities associated with the Western blots shown in several figures. We have further investigated the matter and found that Fig. 2B, 2C, and 3C had been inappropriately assembled. We sincerely apologize to the scientific community for any inconvenience resulting from the publication and subsequent retraction of this article.”
Pubpeer: https://pubpeer.com/publications/2A571C96FF7663023B3DAB3CBE133A
LikeLike
2020 retraction for:
J Biol Chem. 2001 Apr 13;276(15):12068-75. doi: 10.1074/jbc.M006462200. Epub 2001 Jan 11.
Selective cleavage of BLM, the bloom syndrome protein, during apoptotic cell death
O Bischof 1, S Galande, F Farzaneh, T Kohwi-Shigematsu, J Campisi
Affiliation
1Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley California 94720, USA.
PMID: 11154689
4 comments on PubPeer (by: Scopula Submutata, Psoralea Filifolia, Cricotopus Wangi, Streptomyces Melanogenes)
DOI: 10.1074/jbc.M006462200
2020 retraction notice.
https://www.jbc.org/content/295/49/16905
VOLUME 276 (2001) PAGES 12068–12075
This article has been withdrawn by the authors except Dr. Kohwi-Shigematsu, who could not be reached. Fig. 3B has a duplication of the top band in lanes 1 and 3. Fig. 5D has a duplication between the top bands and a horizontal flip of the bottom bands. Fig. 5E likewise has a duplication of the top bands excluding the probe lanes and a horizontal flip of the bottom bands. Fig. 6A has smaller microscopy images pasted on the background of larger microscopy images.
LikeLike
Big shit (american slang/vulgar/sarcasm for “its not important”). Oliver’s plan worked: he worked for someone famous (Campisi–because she comes off well in TED talks and is good at shilling (american slang/vulgar for “self promotion”) senescent cells), published “stuff” (albeit with a little massaging) in high impact factor journals, and is running a lab at a preeminent institute in the shadow of our Dark Lord of Apoptosis Koemer. He is not a failed scientist! You’re wasting your time, Zeb. Oliver is there to stay, especially if he is funded off European grant money and makes his lab personnel wear shilling T shirts.
LikeLike
it’s a laugh though.
LikeLike
Senolytic shilling.Give it up, Judy.Just stop it.
LikeLike
In reply to:
NMH, the failed scientist and incel
December 4, 2020
” Oliver is there to stay, especially if he is funded off European grant money and makes his lab personnel wear shilling T shirts.”
The reality supports what you write.
Two examples from prominent U.S. medical schools:-
1. Albert Einstein College of Medicine, Bronx, New York.
https://retractionwatch.com/2015/09/16/fourth-retraction-for-einstein-oncologist-due-to-image-manipulations/
Yet in 2020 we find: https://www.einstein.yu.edu/faculty/8581/roman-perez-soler/
as if nothing happened.
Weill Cornell College of Medicine, Manhattan, New York.
https://retractionwatch.com/2020/01/03/prominent-cancer-researcher-loses-eight-papers-making-11/
Yet we find: https://research.cornell.edu/researchers/andrew-j-dannenberg
as if nothing happened.
LikeLike
Continuation 2020 retraction for:
J Biol Chem. 2001 Apr 13;276(15):12068-75. doi: 10.1074/jbc.M006462200. Epub 2001 Jan 11.
Selective cleavage of BLM, the bloom syndrome protein, during apoptotic cell death
O Bischof 1, S Galande, F Farzaneh, T Kohwi-Shigematsu, J Campisi
Affiliation
1Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley California 94720, USA.
PMID: 11154689
From the retraction notice.https://www.jbc.org/content/295/49/16905.full
“This article has been withdrawn by the authors except Dr. Kohwi-Shigematsu, who could not be reached.”
I think she is at UCSF.https://profiles.ucsf.edu/terumi.kohwi-shigematsu
Terumi Kohwi-Shigematsu, PhD
Title(s) Professor, Orofacial Sciences
School School of Dentistry
Address 513 Parnassus Ave, HSW
San Francisco CA 94143
Phone 415-502-7828
Email Terumi.Kohwi-Shigematsu@ucsf.edu
vCard Download vCard
Dr. Kohwi-Shigematsu lists it as her publication number 46.
Selective cleavage of BLM, the bloom syndrome protein, during apoptotic cell death. J Biol Chem. 2001 Apr 13; 276(15):12068-75. Bischof O, Galande S, Farzaneh F, Kohwi-Shigematsu T, Campisi J. PMID: 11154689.
https://bms.ucsf.edu/people/terumi-kohwi-shigematsu-phd
In the phone book even.
Terumi Kohwi-Shigematsu, PhD
Professor in Residence
Orofacial SciencesTerumi.Kohwi-Shigematsu@ucsf.edu
+1 415 502-7828
LikeLike
Way too much work… so tired. /s
LikeLike
J Biol Chem. 1999 Jul 16;274(29):20521-8. doi: 10.1074/jbc.274.29.20521.
Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences
S Galande 1, T Kohwi-Shigematsu
Affiliation1Life Sciences Division, Lawrence Berkeley National
Laboratory, University of California, Berkeley, California 94720, USA.PMID: 10400681 DOI: 10.1074/jbc.274.29.20521
Problematic data figure 1B. Much more similar than expected.
LikeLike
Additional problematic data J Biol Chem. 1999 Jul 16;274(29):20521-8. doi: 10.1074/jbc.274.29.20521.
Figure 1B continued. Much more similar than expected.
LikeLike