Who are those people running papermills? Well, Smut Clyde managed to find one owner, a young Chinese gentleman named Chengcao Sun from Wuhan. Sun published oodles of research papers and sold even more, all massively referencing him. This young professor, celebrated in China as scientific prodigy, is not only a successful papermill entrepreneur, but will soon become a Highly Cited Researcher!
Because scholarly publishing is completely broken. We have the usual suspects: Aging, Oncotarget, and a certain Elsevier journal named “Gene“.
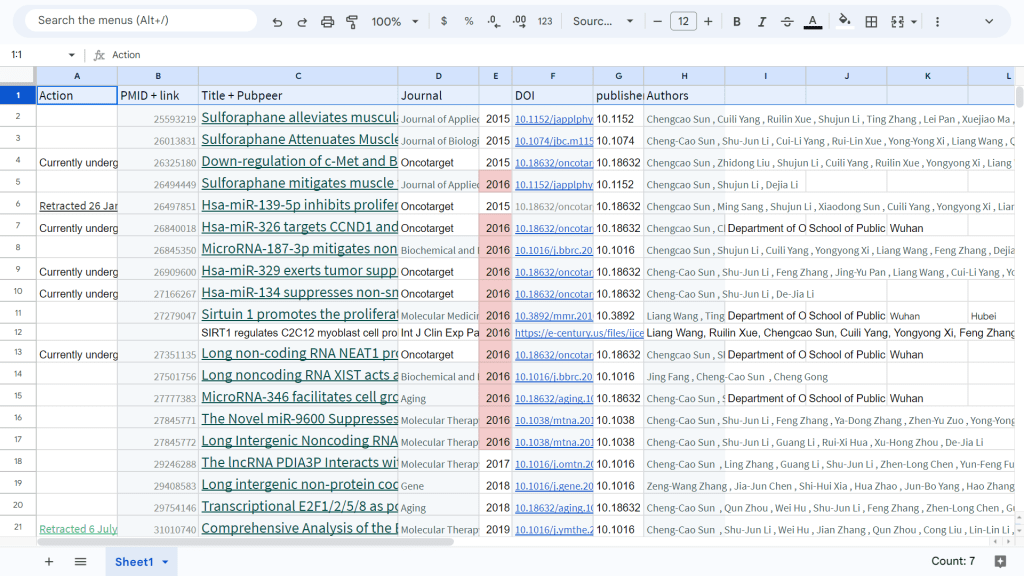
Smut Clyde of course made a spreadsheet, with over 40 entries, here it is:
And now….

Ash Ra Template
By Smut Clyde
 Our host hinted the other day about the importance of templates in science publishing. For each specialty within the biomedical literature there are conventions of the genre – the form that an incremental fingernail-clipping of a study should take, and the kinds of experiments and quantifications required to test the hypothesis, and the set sequence for the Introduction and Discussion to follow.
Our host hinted the other day about the importance of templates in science publishing. For each specialty within the biomedical literature there are conventions of the genre – the form that an incremental fingernail-clipping of a study should take, and the kinds of experiments and quantifications required to test the hypothesis, and the set sequence for the Introduction and Discussion to follow.
Ticket to Tianjin
“So here is a novelty in the annals of fictional research: a nomadic digital caliper. It visited a series of laboratories, accompanied by a backing troupe of mouse-mined xenograft tumours for it to measure” – Smut Clyde
If a manuscript departs from these conventions, the journal editors and reviewers have to think about the purpose and implications of each experiment, much as if they ordered Romance novels from the library but received Westerns instead. There will be toys flung and tears before bedtime. Reviewer 3 asks for further experiments of a more familiar nature, or the editor suggests referring the manuscript to some more appropriate journal. Conversely, if their expectations are met then they can hit the Accept button without needing to scrutinise the Figures or Tables. Following a template makes life easier for everyone.
This is an issue for current attempts to train a Machine Learning model to filter the papermill products from legitimate papers (or at least to assign manuscripts a ‘papermill’ score, to alert editors when to heighten their vigilance). The initial results are promising… relative to the wider biomedical literature, junk papers do seem to be concentrated in specific zones of maps from (e.g.) the “Clear Skies” detector, or “The Landscape of Biomedical Research” (Actinopolyspora biskrensis regularly credit the Clear Skies detector in PubPeer comments). To put it another way, certain regions of ‘paper space’ are hotspots of papermill activity.

[right] González-Márquez et al 2023.
The ‘null hypothesis’ is that (1) these hotspots are simply regions of heightened activity, where all the cool kids are reporting results, meaning that wannabee authors want to publish there too, while publishers are especially eager for material to publish; and (2) collapsing a high-dimensional space into 2D has actually produced a detector for templates – which are common in papermill products but not confined to them. I remain unconvinced that there are universal distinguishing traits in the structural and linguistic stylistics of all papermill output that a model could be trained to detect.
Setting that aside, sometimes the templating becomes too prescriptive and bedecks a text with curious, idiosyncratic, easily-searchable turns of phrase:

Closer inspection of this little genre reveals Introductory and Discussion passages that follow a stylised course. Clearly a number of papers have been hastily assembled from Lego-blocks of prose, hewing closely to a template – one that seems to have been tailored for the Elsevier journal Gene, where most of the manuscripts found homes. All on the general topic of long non-coding RNAs as facilitators or markers of (or impediments to) cancer. Here are some more of the verbal building-blocks:

Another observation follows! All the teams of researchers hail from hospitals in Wuhan, attached to Wuhan Medical University.1 Some were repeat authors:
- From the Department of Otorhinolaryngology-Head and Neck Surgery, Zhongnan Hospital: Peng Song, Tao Peng, Xu-Hong Zhou and others have their names on six papers, with an emphasis on curing nasopharyngeal carcinoma.
- From the Department of Radiation and Medical Oncology, Zhongnan Hospital: Gang Chen and others signed one monograph on lung cancer and another on nasopharyngeal carcinoma.
- From Wuhan University’s School & Hospital of Stomatology: Chen-Zheng Zhang was sole author of two monographs on oral squamous-cell carcinoma.

Following on from the stilted phrases, the template includes slots for colony formation plates, Western Blots, and immunostaining figures. These were filled by repurposing images across the entire corpus of papers, which is not ideal, but reviewers were lulled into slumber by the familiar comforting cadences of the prose.
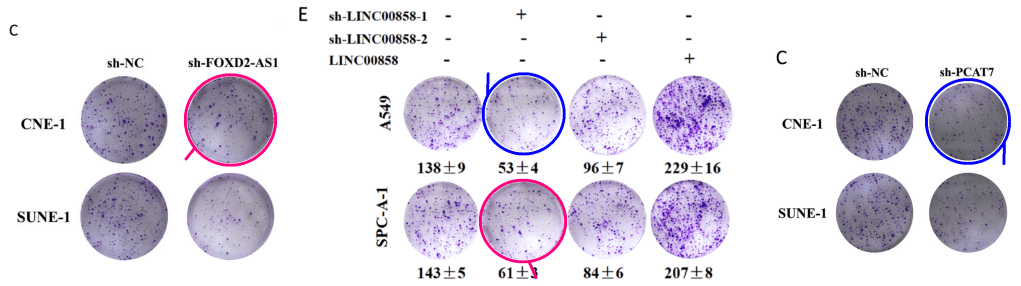
Fig 2E from “Long intergenic non-protein coding RNA 00858 functions as a competing endogenous RNA for miR-422a to facilitate the cell growth in non-small cell lung cancer” (Zhu et al 2017).
Fig 2C from “Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma” (Liu et al 2017)..

[right] Fig 2C from “Long non-coding RNA 520 is a negative prognostic biomarker and exhibits pro-oncogenic function in nasopharyngeal carcinoma carcinogenesis through regulation of miR-26b-3p/USP39 axis” (Xie et al 2019).


About half this corpus was co-authored by Cheng-Cao Sun, often collaborating with De-Jia Li (whom, I intuit, was Sun’s mentor), both of Wuhan University’s School of Public Health.2 My spreadsheet includes Sun’s 2015 papers on curing muscular dystrophy with sulforaphane (i.e., cabbage squeezings), for they are riddled with recycled and manipulated microphotographs of Mus musculus muscle tissue.

[right] Fig 3A from “Sulforaphane Attenuates Muscle Inflammation in Dystrophin-deficient mdx Mice via NF-E2-related Factor 2 (Nrf2)-mediated Inhibition of NF-κB Signaling Pathway” (Sun et a 2015b).
But the story really gets underway in 2016. This was an annus mirabilis for Sun, with 12 papers published.
Sun was still a PhD student in 2016, while winning prize after prize for the quality of his research, with a bright future officially foreseen for him. Seemingly he had been selected for a meteoric, fast-track ascent.

After a post-doctorate fellowship at the MD Anderson Cancer Center at University of Texas, our man is already an Assistant Professor, with the attendant responsibilities of journal Editorial Boards and Special-Issue Guest Editor. His productivity dropped from 2018 onwards, though COVID created openings for a few papers on its treatment, and there was time for glowing media coverage of his lung-cancer-treatment breakthroughs.
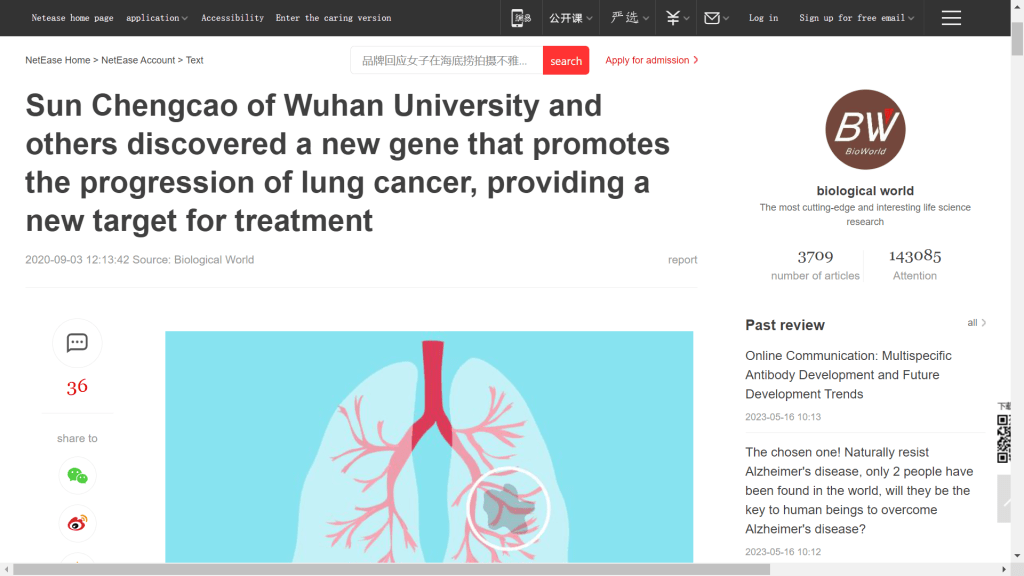
Sun’s coverage from Retraction Alert / News was slightly less laudatory. This being a Chines-language research-integrity blog carried under the auspices of Zhihu.com, though picked up and amplified by other aggregation sites.
“Recently, a number of SCI papers published by Wuhan University around 2016 have been questioned on Pubpeer for improper use of pictures, suspected fraud and many other issues that have attracted attention.”
Ruler of the Aging Papermill
Smut Clyde congratulates Aging: “This is bespoke tailoring, in contrast to the off-the-rack products cranked out by the average papermill […] no shame befalls the journals that accept these confections.”
This happened in late 2022 when even the Editors of Oncotarget had reluctantly noticed the industrial-scale recycling of colony-plate images, and the “Under Investigation’ flags proliferated like a mutant form of measles. Which is more than Elsevier journals have achieved. Meanwhile a 2019 Sun paper was depublished from Molecular Therapy.

The anonymous Retraction News journalists read PubPeer, and cheerfully summarised what was known by then in a table (duly stolen, above). They speculated about a possible papermill contribution to Sun’s productivity.
“There are indications that these eight highly similar articles have traces of “mass production” and may have come from “paper factories“ .
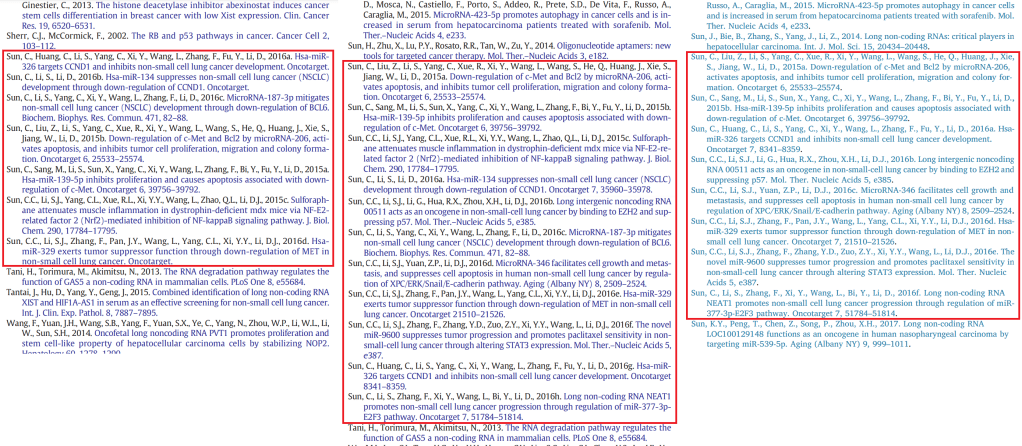

There is another possibility, though… that C.-C. Sun is the papermill, supplying colleagues around Wuhan with variants of the same paper, with the condition that those donated manuscripts carry self-citation payloads. For another feature of the template is an inordinate fondness for the oeuvre of C.-C. Sun, whose doubtless-seminal papers – including review papers, and the 2015 cabbage-squeezing research – are cited in great slabs in the Introductory statements about the prevalence of cancers (where some primary sources might be more appropriate), and for the details of various experimental procedures. One could easily infer that no other authorities exist in this field of long-non-coding RNAs.

One author used the name of another author to leave a comment at PubPeer, explaining that the Sun-centric nature of the References merely reflected Dr Sun’s unacknowledged role in preparing the manuscript.
#2 Yong-Shun Chen commented May 2023
Dear Hoya camphorifolia : Of course, many references from a single author is unusual for a paper. However, the references of the Introduction Section and Methods section of the article, are not randomly selected. First of all, Dr Sun Chen-cao gave us valuable suggestions on the experiment section, some of which are based on his previous publications. Secondly, the references fit the context of our article, and these references will make our manuscript more logical and reasonable. Last but not least, these references does not affect the article’s fundamental conclusion. Thank you very much for the problems you pointed out. In our following manuscripts, we will be more cautious to avoid this situation. Best wishes, and your sincerely, Ke Shaobo.
All this is certainly one way to raise the profile of one’s research and ensure that it is widely cited. I am impressed, though, that Sun found any time at all to conduct actual experiments.
The papermills of my mind
“For as I have often bemoaned in the past, not even the paper-forging industry is free from scruple- and principle-deficient players.” – Smut Clyde
I promised Western Blots and immunostaining figures, so here they are:

[right] Fig 9B from “MicroRNA‐346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non‐small cell lung cancer by regulation of XPC/ERK/Snail/E‐cadherin pathway” (Sun et al 2016).

Fig 2F from “LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6” (Kong et al 2018).
Fig 4E from “Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p” (Sun et al 2017).

[below] Fig 4E from “Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p” (Sun et al 2017).

Notes
1. There are two exceptions to this “All from Wuhan” generalisation.
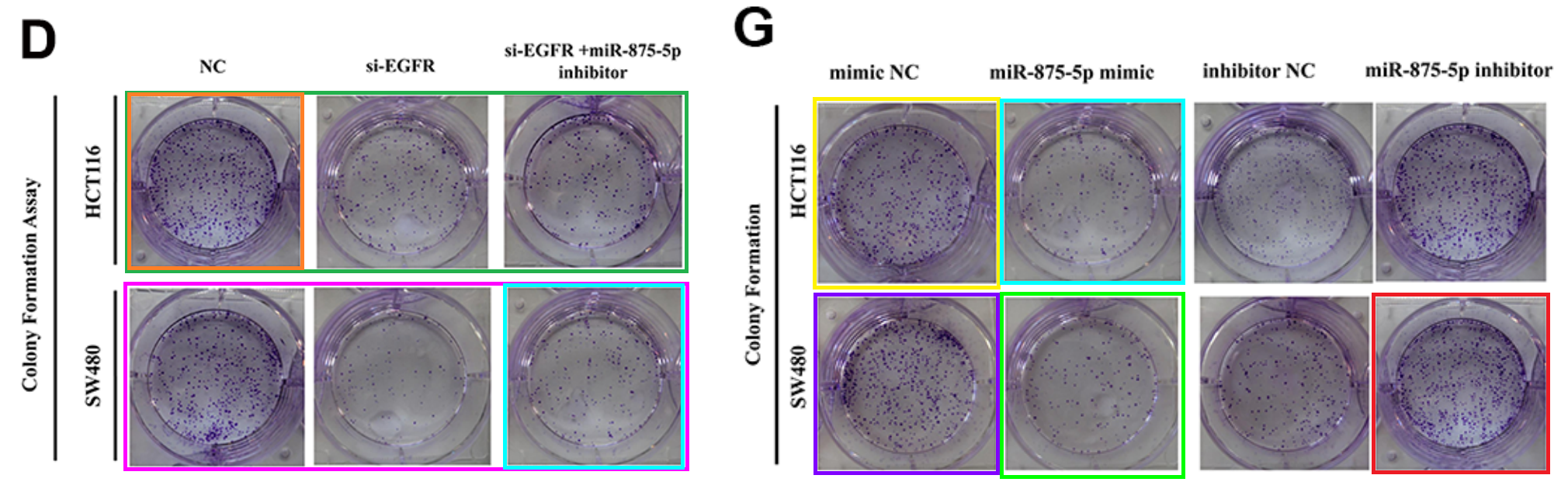
- “Hsa-miR-875-5p exerts tumor suppressor function through down-regulation of EGFR in colorectal carcinoma (CRC)” (Zhang et al 2016) was signed by oncologists at Shanghai Jiaotong University, Medical College.
- “CircDUSP16 promotes the tumorigenesis and invasion of gastric cancer by sponging miR-145-5p” (Zhang et al 2020), from Shanghai Ren Ji Hospital, is something of an outlier with a total absence of Sun references. Although it came right at the end of this sequence of papers, the lead author reports that the research had been in progress since 2014, giving them a prior claim to the loading band that also features in two 2016 papers from the School of Sun.

Fig 3F from “CircDUSP16 promotes the tumorigenesis and invasion of gastric cancer by sponging miR-145-5p” (Zhang et al 2020).
Fig 8D from “Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1” (Sun et al 2016).
An absence of Sun references is also what convinced me to leave “Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer” (Shi et al 2016) out of the spreadsheet.

Fluorescing-cell panels from Figs 3G-I and 7E,F,I of that paper do appear in Fig 3C,D of “Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p” (K.-Y. Sun et al 2017); and in Fig 2G of “FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway” (C.-C. Sun et al 2020). However, Shi et al (with Nanjing affiliations) appear to be innocent donors of the shared material.
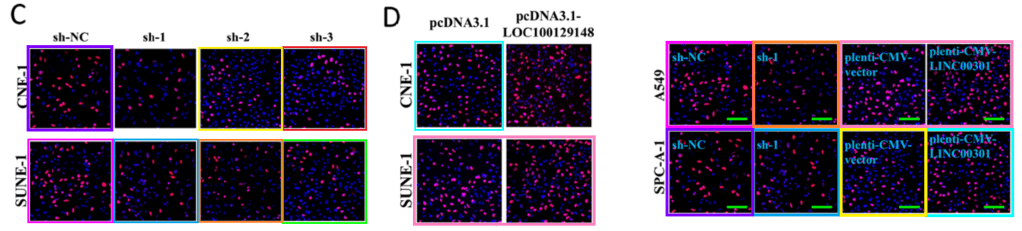
Right: Sun et al 2020.
This curious backstory to Sun et al (2020) was not covered by the 2021 correction of a tiresome panel duplication and a faulty mouse-breed identification.

Nor did it affect the promotion of Sun et al (2020) by “Wuhan University News Network”, gratefully regurged by churnalism sites.
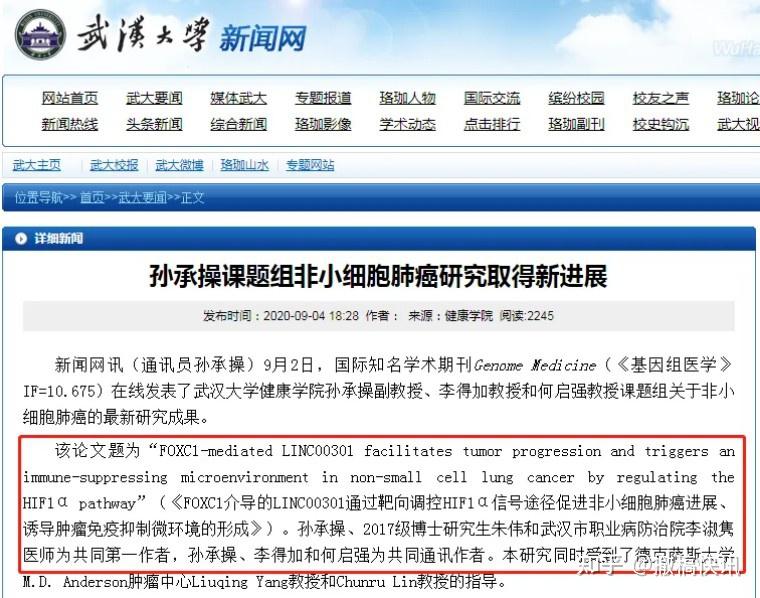


2. Coincidentally, Professor De-Jia Li is on the Editorial Board at Gene.
Donate to Smut Clyde!
If you liked Smut Clyde’s work, you can leave here a small tip of 10 NZD (USD 7). Or several of small tips, just increase the amount as you like (2x=NZD 20; 5x=NZD 50). Your donation will go straight to Smut Clyde’s beer fund.
NZ$10.00





“All the teams of researchers hail from hospitals in Wuhan, attached to Wuhan Medical University”
There’s so much outstanding research in lovely Wuhan. miRNAs, cancer, bat coronaviruses… I really feel reassured by the tireless work of these great scientists who are committed to a better world.
LikeLike