Scholarly publishing: another sad case where giant industrial companies take over the market and push aside small and medium-sized artisan businesses. Hindawi, MDPI, Spandidos: they all chose Big Papermill multinational conglomerates as their business partners, but who will spare a thought for the local cottage papermills, where every customer is greeted personally, and whose travelling salesmen drop in for tea?
Luckily, there are still journals who open their doors to such salesmen, because they prefer locally-sourced custom-made papermill fraud to anonymous mass-produced fraud of the Big Papermill industry. Thank you, Frontiers, thank you Misha Blagosklonny’s Oncotarget and Aging, thank you Springer Nature’s Cell Death & Disease and Scientific Reports, thank you all those other Open Access journals. Where would the small but fine Jammy Smears Papermill be without you? Selling essays to undergrad students at the local college probably.
This new revolutionary investigation by Smut Clyde wants you to appreciate the little things. Follow Smut to the garbage dump of scholarly publishing.
The papermills of my mind
By Smut Clyde
Please admire this purchased manuscript, elevated to the giddy empyrean of Cell Death & Disease‘s 2019-2021 “Readers’ Choice” list of Outstanding Papers, with plagiarised illustrations and 89 citations to date (by the inflated count of Goofle Scholar). Its apotheosis is a tribute to the anonymous papermill authors, who evidently hit the right notes and know exactly what appeals to CDD readers (though it does not redound to the credit of that readership, nor to the CDD editors and reviewers).
Cell Death and Depravity
Is the journal Cell Death and Disease a disease itself, parasitised by Chinese paper mills? Can it be cured? Not with this team of doctors on editorial board.
It appears that exosomes are a permanent addition to papermill praxis, and that the interplay between non-coding RNAs and cell-signal pathways is no longer enough to create a truly satisfying title.


The elevation also makes up for three other manuscripts, signed by the same customer and subsequently consigned to the ignominy of retraction.



In fact there is a cluster of 22 papers (so far) with implications for cancer treatment, linked by shared or overlapping figures. Six comprise the CV of Huaying Dong and colleagues from Hainan General Hospital, Jinan University, but most have unrelated authors.

In the course of writing mumblety-mumble guest blogposts at For Better Science I have focused on and sought out prolific papermills… drawn by the satisfaction that comes from giving a local habitation and a name to a large group of fake papers, tying up 700 or 1000 of them at once. I have also written about the junkpaper industry as if it consisted of three-way transactions between (1) the papermills, wholesale producers of falsity; (2) the purchasers who sign the manuscripts; and (3) the final consumers, the journals – with the machinery of nescience kept running by the flow of funding from (2) to (1) and (3).
The full-service paper mill and its Chinese customers
An investigation by Elisabeth Bik, Smut Clyde, Morty and Tiger BB8 reveals the workings of a paper mill. Its customers are Chinese doctors desperate for promotion. Apparently even journal editors are part of the scam, publishing fraudulent made-up science.
“We are extremely guilty and distressed”
“Now no-one wants ForBetterScience to become an all-Papermill channel. And we cannot really expect to shame or inspire scriveners in the academic-ghostwriter industry to seek out more constructive applications for their talents, so the point of exposing them is not immediately obvious.” -Smut Clyde
This is an oversimplified picture, though, as (1) subsumes two different skill-sets and activities. It’s like conflating authors with their agents. Operating the looms of wordwooze assembly is distinct from what I call the ‘dragoman’ role: recruiting the signatories (2) and matching their needs to the house-styles and specialised capabilities of producers, while arranging for complaisant guest-editors and peer reviewers liaising with the journal editors of (3). That is, the ecosystem includes a retail tier as well as the wholesalers; a kind of brokerage. This meshes with Adam Day’s analysis, and with ‘Parashorea tomentella‘s observations of the Hindawi business model where brokers actively tout for would-be researchers who can add their signatures to manuscripts from the brokers’ suppliers. The retail tier allows cottage-industry papermills to stay viable, by tapping into larger distribution networks.
The Rise of the Papermills
“Is it possible that through no fault of Zintzaras & Ioannidis, their work was incorporated into a papermill template, accruing hundreds of spurious citations?” – Smut Clyde
Papermill – your local partner for Special Issues in China
Did you know that special issues are better than regular issues and better than special issues?
A corollary follows (as corollaries are wont to do). Once a distinguishing feature of some style of fabrication has been identified, the possibility remains that it’s the hallmark of a broker, rather than of a specific papermill.
Victims as perpetrators
Smut Clyde as Detective Columbo investigates: The victims of a paper mill are actually in cahoots with the perpetrators! Stealth corrections happen faster than one can catch them!
An illustration is on hand! Recall the ‘hipster herbalism’ papermill that infiltrated and took over J.BUOn, the Journal of the Balkan Union of Oncology (until that journal succumbed to the drain on its credibility), ringing the combinatorial changes on the intersection between varieties of cancer, the menagerie of ncRNAs, and phytochemicals. The miller’s products were crude to start with but they learned quickly to mask the plagiarism and the manipulations. They were especially fond of green-yellow-orange fluorescence microscopy – the spoor of the Horsemen of the Apoptocalypse.
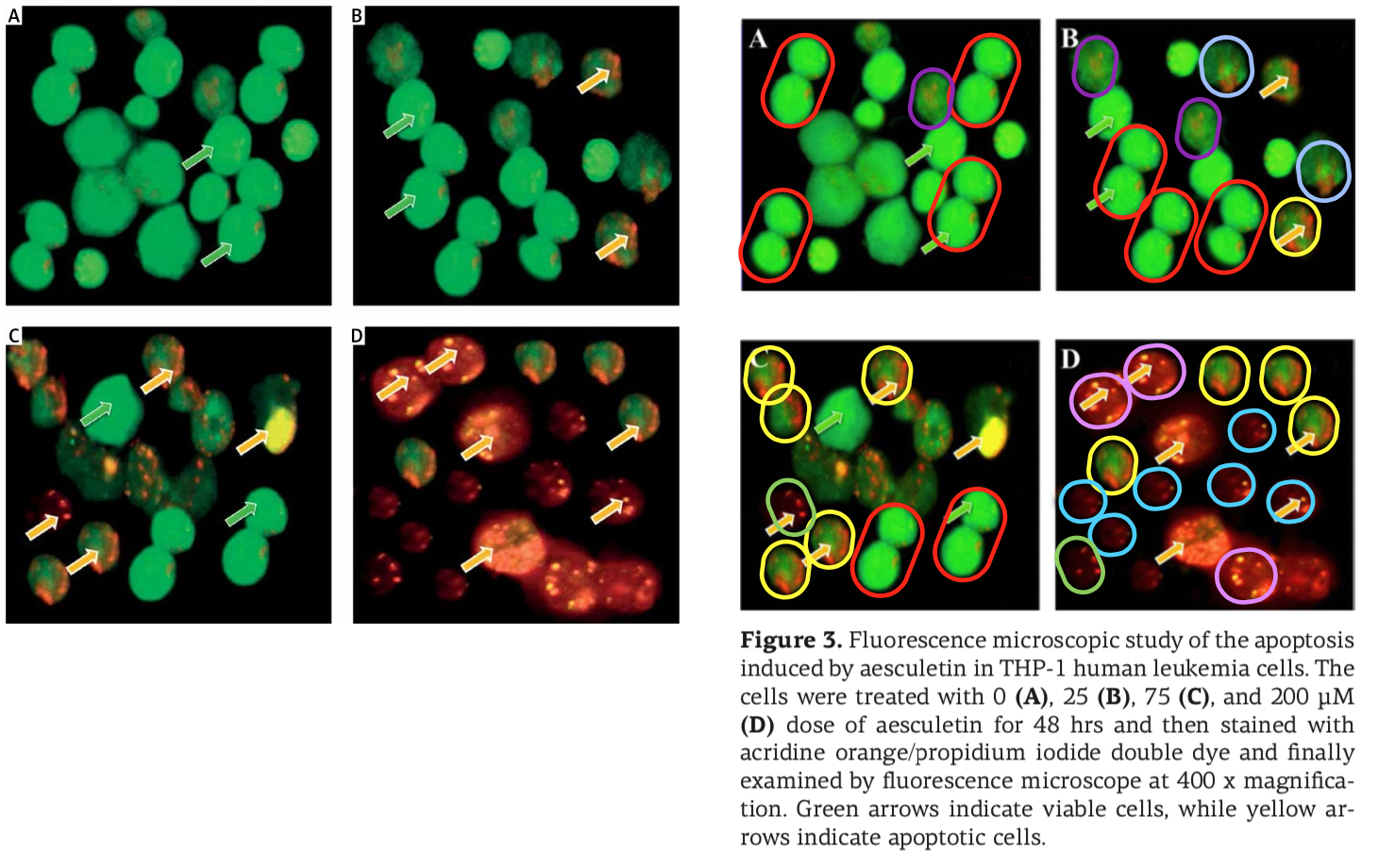
The millers were not tied exclusively to J.BUOn, channeling some of their output through a spectrum of negotiable standards: Spandidos organs, the legendary European Review for Medical and Pharmacological Sciences, Archives of Medical Science and Medical Science Monitor (a pair of Polish journals), and a few from the Springer and Elsevier stables. But conversely, the J.BUOn editors felt somehow beholden to the millers (for reasons that remained as unclear as a pint of Old Sheepshagger Baltic Roggenbock), and they tried to cover up for the malfeasance by stealth-correcting the bogus papers… which often required repeat rounds of correction when the first attempts just introduced new photoshoppery.
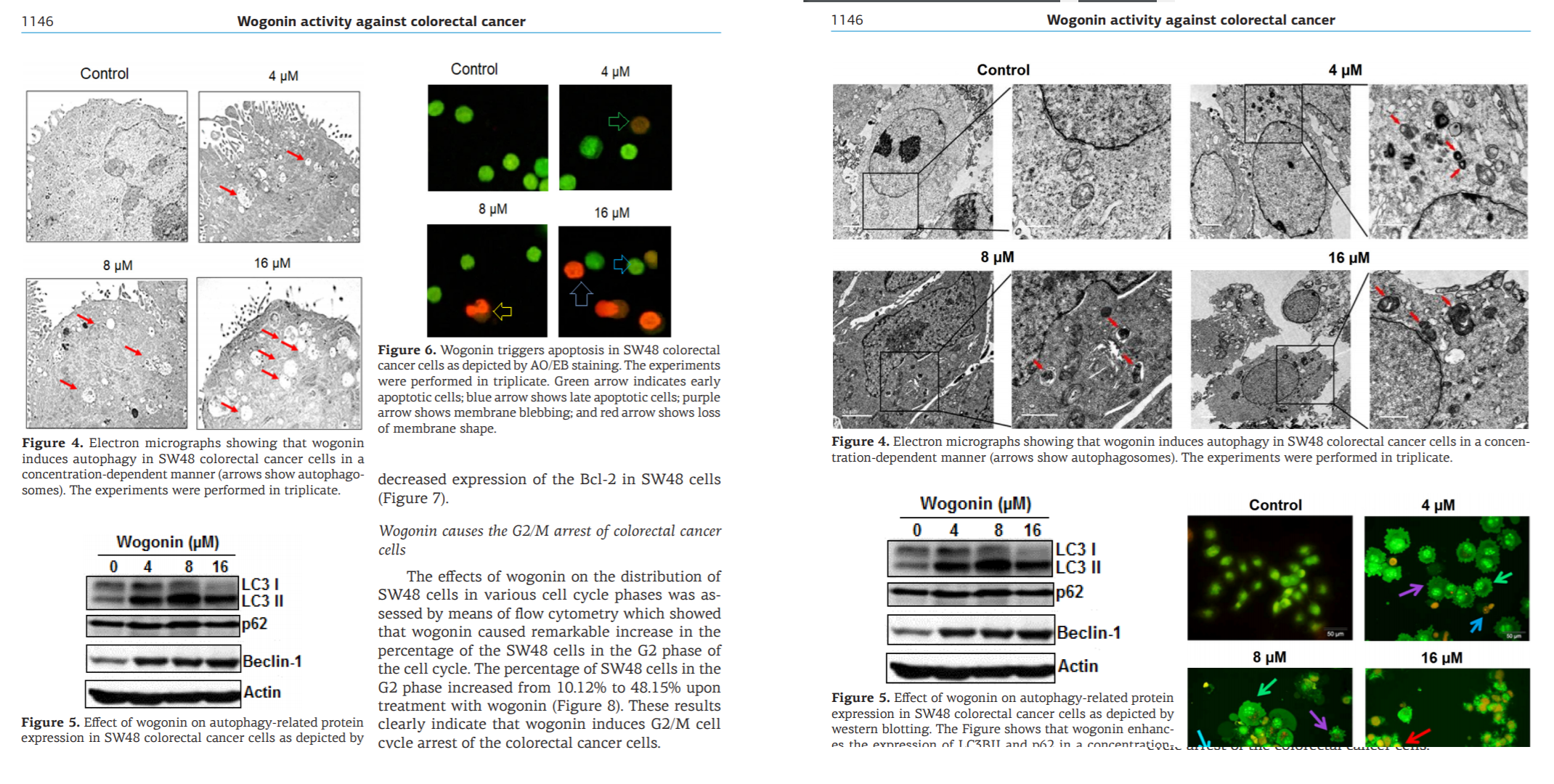
One obvious marker of provenance was the Corresponding Authorship. A gallery of evanescent identities with disposable single-use Yahoo accounts sheltered the nominal, signatory authors from the demands of dealing with author / editor negotiation. Each bizarre name cried out to become the protagonist of a short story in the hard-boiled-detective genre.
TBarnebesaull@yahoo.com sporusac@yahoo.com RichieWattsfdv@yahoo.com BrattinOdod@yahoo.com RLakeshiattebe@yahoo.com StaciNcrod@yahoo.com MMadalynywa@yahoo.com AlistadLhos@yahoo.com DCassienne@yahoo.com KNanceylse@yahoo.com DanyelSsche@yahoo.com RBellengob@yahoo.com
The “Jammy Smears” papermill
 But an equally implausible dramatis personae features as the corresponding authorship of a second, stylistically-different oeuvre. For convenience I’m calling this the ‘Jammy Smears‘ papermill [link to spreadsheet], after the distinctive Western Blots where a limited alphabet of smeary blobs are shuffled with minimal effort into the equivalent of ransom notes. If some Victorian entomologist compiled collections of insect larvae by pressing them flat in an album, in the manner of flowers and fairies, the results would be similar.
But an equally implausible dramatis personae features as the corresponding authorship of a second, stylistically-different oeuvre. For convenience I’m calling this the ‘Jammy Smears‘ papermill [link to spreadsheet], after the distinctive Western Blots where a limited alphabet of smeary blobs are shuffled with minimal effort into the equivalent of ransom notes. If some Victorian entomologist compiled collections of insect larvae by pressing them flat in an album, in the manner of flowers and fairies, the results would be similar.

[middle] Fig 5 from “Pristimerin inhibits neuronal inflammation and protects cognitive function in mice with sepsis-induced brain injuries by regulating PI3K/Akt signalling” (Xue et al 2021).
[right] Fig 4A from “Fuscoside Attenuates Bone Loss in Bone Defects by Regulating The Rankl/Nlrp3/Opg Pathway in Rats” (Liu & Wang 2021)

[upper right] Fig 5 from “Fuscoside Attenuates Bone Loss in Bone Defects by Regulating The Rankl/Nlrp3/Opg Pathway in Rats” (Liu & Wang 2021).
[below] Figs 3a, 6a from “Desoxyrhapontigenin attenuates neuronal apoptosis in an isoflurane-induced neuronal injury model by modulating the TLR-4/cyclin B1/Sirt-1 pathway” (Liang et al 2020).

“Jammy Smears” covers similar subject matter to “Hipster Herbalism” – random secondary plant metabolites, treating random conditions by modulating random signal pathways – and reasonably enough it targets the same journals. My working hypothesis is that these ephemeral Yahoo accounts were all spawned by a single broker who was recruiting signatories and lining up publishers for two separate studios.
 In this case the Acknowledgements seem to be contributed by the second papermill. They are typically boilerplate and fulsome, thanking the authors’ institutions for providing the “necessary facilities” that made the fantasy research possible. In contrast, searching Goofle Scholar or Dimensions for email identities of the form “*@yahoo.com” leads quickly into both oeuvres.
In this case the Acknowledgements seem to be contributed by the second papermill. They are typically boilerplate and fulsome, thanking the authors’ institutions for providing the “necessary facilities” that made the fantasy research possible. In contrast, searching Goofle Scholar or Dimensions for email identities of the form “*@yahoo.com” leads quickly into both oeuvres.

So welcome to the world of small-batch, artisanal, cottage-industry papermilling; the Mittelstand of parascience fabrication, as it were. Other mills in this class lack such obvious flags so there is no telling what proportion of each studio’s production has so far been detected, nor how many are still out there. The “Cangzhou / Aging” mill seems to fit into this tier.
Ruler of the Aging Papermill
Smut Clyde congratulates Aging: “This is bespoke tailoring, in contrast to the off-the-rack products cranked out by the average papermill […] no shame befalls the journals that accept these confections.”
The Adventures of a Mouse Malignancy Group Portraitist
Smut Clyde was busy with yet another Chinese paper mill. A plastic ruler was deployed.
All we have is a small cluster or network of papers from each studio, linked to one another by a shared design template, and by overlapping images from some experimental procedure – Transwell assays, or Western Blots, or flow cytometry (it goes without saying that beta-tester access to ImageTwin facilitates this work). Even though these pictorial data are often manipulated as well as overlapping, they do seem to be generally grounded in reality (excepting the jammy smears just described), making them less obvious than algorithm-generated WBs in the styles of ‘tadpoles’, or processionary caterpillars, or strings of sausages or bowties. Nor is there any telling how many of these fake-paper clusters exist, but it might be time to revise estimates of the frequency of forgery (especially in the biotech literature, in trending topics like non-coding RNAs or exosomes or both).

The “All patients” papermill
Time for another example? The “All Patients” cottage corpus [link to spreadsheet] was recognised and catalogued by Elisabeth Bik.
Recycled and sometimes retouched Transwell Invasion / Migration images distinguish the mill’s products, though Western Blots recur as well when those were what the journal preferred.
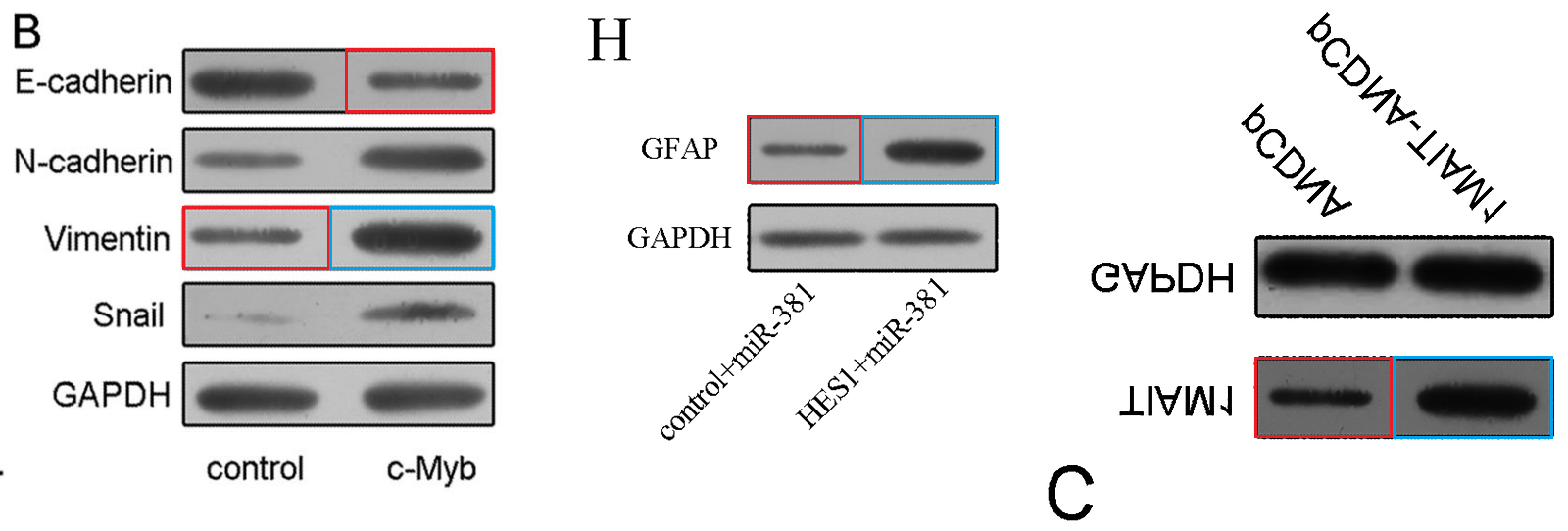
Fig 6H from “miR-381 Regulates Neural Stem Cell Proliferation and Differentiation via Regulating Hes1 Expression” (Shi et al, 2015).
Fig 5C from “The Downregulation of MiR-182 Is Associated with the Growth and Invasion of Osteosarcoma Cells through the Regulation of TIAM1 Expression” (Hu et al, 2015). Flipped vertically for convenience.
Dr Bik’s title “All Patients” reflects the millers’ predilection for boilerplate Ethics Declarations in which “All patients agreed to participate”. I would just like to announce, by the way, my own policy of naming newly-characterised papermills after my most generous Patreon donors.
Anyway, this studio was fond of PLoS One as an outlet (occasionally branching out to OncoTarget, and then further afield to Journal of Cellular and Molecular Medicine), though a recent shift in PLoS policy to reduced tolerance for fabrication brought the Scythe of Retraction cutting swathes through that back-catalogue. Instances are still emerging from the woodwork, meaning that my list is substantially embiggened from Dr Bik’s: 56 65 entries so far (nearly half in PLoS One), with 15 retractions.

The mill’s activity peaked about 2014-2016, though later instances were still emerging from the pipeline in 2018/2019. The earliest instances date back to 2013 (h/t ‘Campanula pseudostenocodon‘), and frequently have two authors in common: Fang Wang and Jia Yu from Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & Peking Union Medical College. In fact that pair signed ten twelve of the “All Patients” papers, the latest in 2019 – earning them a customer-loyal discount card, unless they are actually the source. The names of other staff from Institute of Basic Medical Sciences also frequently appear.

Fig 3d from “The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer” (Guo et al 2013).
Fig 4c from “MicroRNA-33b Suppresses Migration and Invasion by Targeting c-Myc in Osteosarcoma Cells” (Xu et al 2014) [retracted].
Fig 7D from “TOX Acts an Oncological Role in Mycosis Fungoides” (Yu et al 2015).
 As Corresponding Author of Li et al (2015), Jia Yu actually responded in one of the Pubpeer threads, explaining that
As Corresponding Author of Li et al (2015), Jia Yu actually responded in one of the Pubpeer threads, explaining that
- The lead author had left the laboratory before the paper was published, taking the raw images, and is responsible for the appearances of the same Figures in papermill products; and
- He was unaware that his email address had been used to submit the paper.
Given that the Corresponding Author had disowned the paper, we can only speculate who submitted the Correction that the journal subsequently accepted.
A 2020 paper in Journal of Cancer uses figures from “All Patients” but I believe Chen et al (2020) to be an independent fraud, from a faker stealing images and showing poor judgement in their sources. For as I have often bemoaned in the past, not even the paper-forging industry is free from scruple- and principle-deficient players.
The papers are coming from inside the house!
“It feels like half the higher-echelon professors at Jilin University have built their careers on these fairy-tales, with successions of papers itemising the interactions of ADAM10 or GRIM-19. […] if only they had published instead about the Tooth-Fairy circ-RNA and how it targets the Easter-Bunny Pathway…”, – Smut Clyde
Spandidos and the Paper Mill
Papermills run by Chinese universities and funding a notorious Greek publisher. Smut Clyde tells it all!
There is no sharp boundary between a ‘papermill’ as a purely commercial endeavour, and a departmental practice of faking data in-house to provide the faculty and grad students with publications, with the resulting corpus augmented by a few fake papers sold on the side to friends and colleagues. The Jilin and CSU (Central South University) institutional papermills are large-scale examples of the latter. The picture is further complicated when outside plagiarists steal image from a papermilled fake: at least one product of the Jilin mill is illustrated with panels ripped off from ‘All Patients’. I am not angry with them, just very disappointed.

In another mill’s oeuvre, a Western Blot purloins one band from a product of the “Tadpole” papermill (the bands marked with blue and orange come from a different source). This is just sad now.


There is not much to say about this ‘Cottage #10’ except that so far 22 papers are known. Its most unifying trait is a single suite of flow-cytometry scatterplots.



[right] Fig 3G from “Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203” (Zhou et al, 2018).

The “Colony Retraction” papermill
A few authors recur in the corpus of this next cottage. Repeat customers, or members of the production team, who can tell? So far 38 instances, almost all in 2018-2020. I call it the ‘Colony Retraction‘ mill, after the recycled colony-assay plates that are the metaphorical rug tying the whole room together.

Fig 2B from “Increased Six1 expression in macrophages promotes hepatocellular carcinoma growth and invasion by regulating MMP‐9” (Zhang et al, 2019).
Fig 8 from “Antiproliferative and Apoptosis Triggering Potential of Paclitaxel-Based Targeted-Lipid Nanoparticles with Enhanced Cellular Internalization by Transferrin Receptors—a Study in Leukemia Cells” (Dai et al, 2018).
Fig 6A/B from “Cytotoxic Compounds from Juglans sinensis Dode Display Anti-Proliferative Activity by Inducing Apoptosis in Human Cancer Cells” (Lee et al, 2015).
Other unifying threads include the usual impedimenta of WBs and fluorescing tissue slices. The latter are starry-night sky-fields, where constellations of Caspase-3 or TUNEL-positive cells stand out against indigo backgrounds of DAPI.
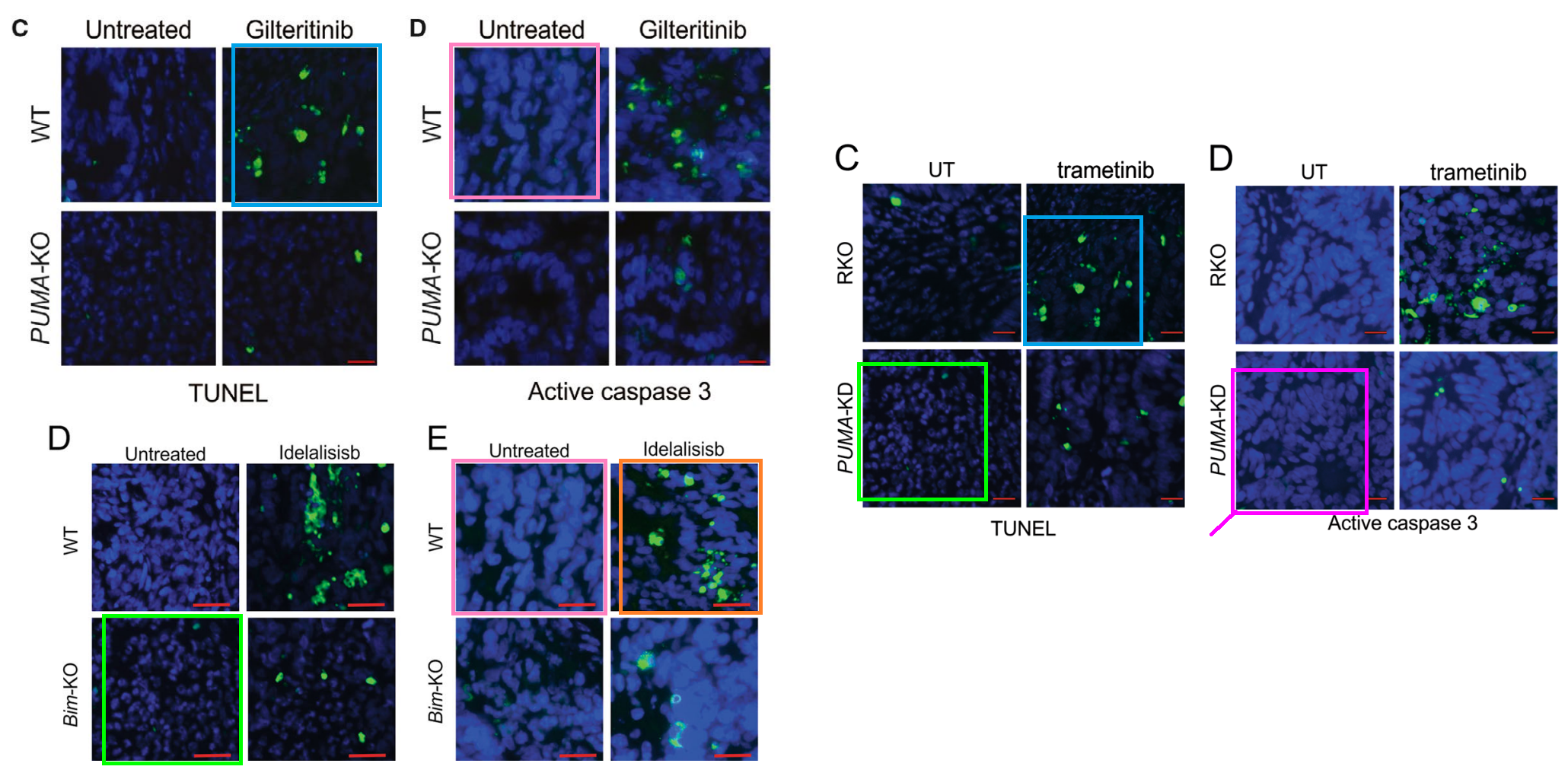

The overlapping protein-stained images of tissue slides were what triggered ‘Actinopolyspora biskrensis’ to flag the papers at the nucleus of the cluster.

[Right] Fig 5D from “The miR-30a-5p/CLCF1 axis regulates sorafenib resistance and aerobic glycolysis in hepatocellular carcinoma” (Zhang et al, 2020).
Authors have been unusually responsive on PubPeer, not providing explanations but at least promising and in a few cases delivering retractions. These responses feature the same wording, again and again, from seemingly unrelated teams from different institutions: “We already submitted retraction letter“, or “We have contacted the journal and required to retract this paper“. All I am saying is that when you realise that the article you published was written by a papermill so you ask the journal to retract it, it would save no end of trouble if you flagged it on PubPeer… rather than waiting for other people to find the recycled / stolen material and then announcing your request.

Here is another montage of Van Gogh-inspired starry nights. Aren’t they restful?

The Shaanxi papermill
I am open to the possibility that as well as recruiting the funders / signatories through advertising in social media, the brokers are also cold-calling genuine laboratories to recruit them as new cottage-industry sources. “Double your funding! Just tweak your published manuscripts with made-up values and out-takes from published figures, and get paid for it!” It would be an especially tempting offer to laboratories where the PI or the pipette jockeys are used to ‘improving’ or cherry-picking their results to ensure that they are sufficiently impactful and publishable.
Those are not the only groups to abandon the quaint, old-fashioned notion that the specimens of experimental output used to illustrate a research narrative should be part of that research. A group of hepatobiliary surgeons at First Affiliated Hospital of Xi’an Jiaotong University have no f***s left to give. Identity parades of xenograft tumours have come unmoored from their origins and can appear anywhere, hugger-mugger and higgledy-piggledy; mere anarchy is unleashed upon the world! Cats and dogs living together! Spreadsheet here.

[right] Figs 8G, 7D from “CXCR4 mediates matrix stiffness-induced downregulation of UBTD1 driving hepatocellular carcinoma progression via YAP signaling pathway” (Yang et al, 2020).

[right] Fig 3A from “KLF10 inhibits cell growth by regulating PTTG1 in multiple myeloma under the regulation of microRNA-106b-5p” (Zhou et al, 2020).

Along with the usual flow cytometry, IHC slides and Transwell invasion / migration assays. This is what happens when you fill the illustration-shaped gaps in a manuscript with random rectangles of wallpaper cut from larger images: overlaps between the pictorial contents of what are supposed to be quite different studies. The originals come from a portfolio of Transwell assays.

[right] Fig 3B,D (Invasion panels) from “Fibulin-5 inhibits hepatocellular carcinoma cell migration and invasion by down-regulating matrix metalloproteinase-7 expression” (Tu et al, 2014b)..
It is probably time for some scratch-assay migration results…
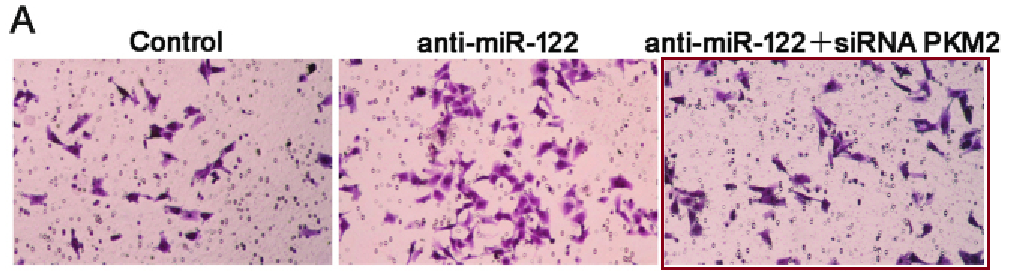
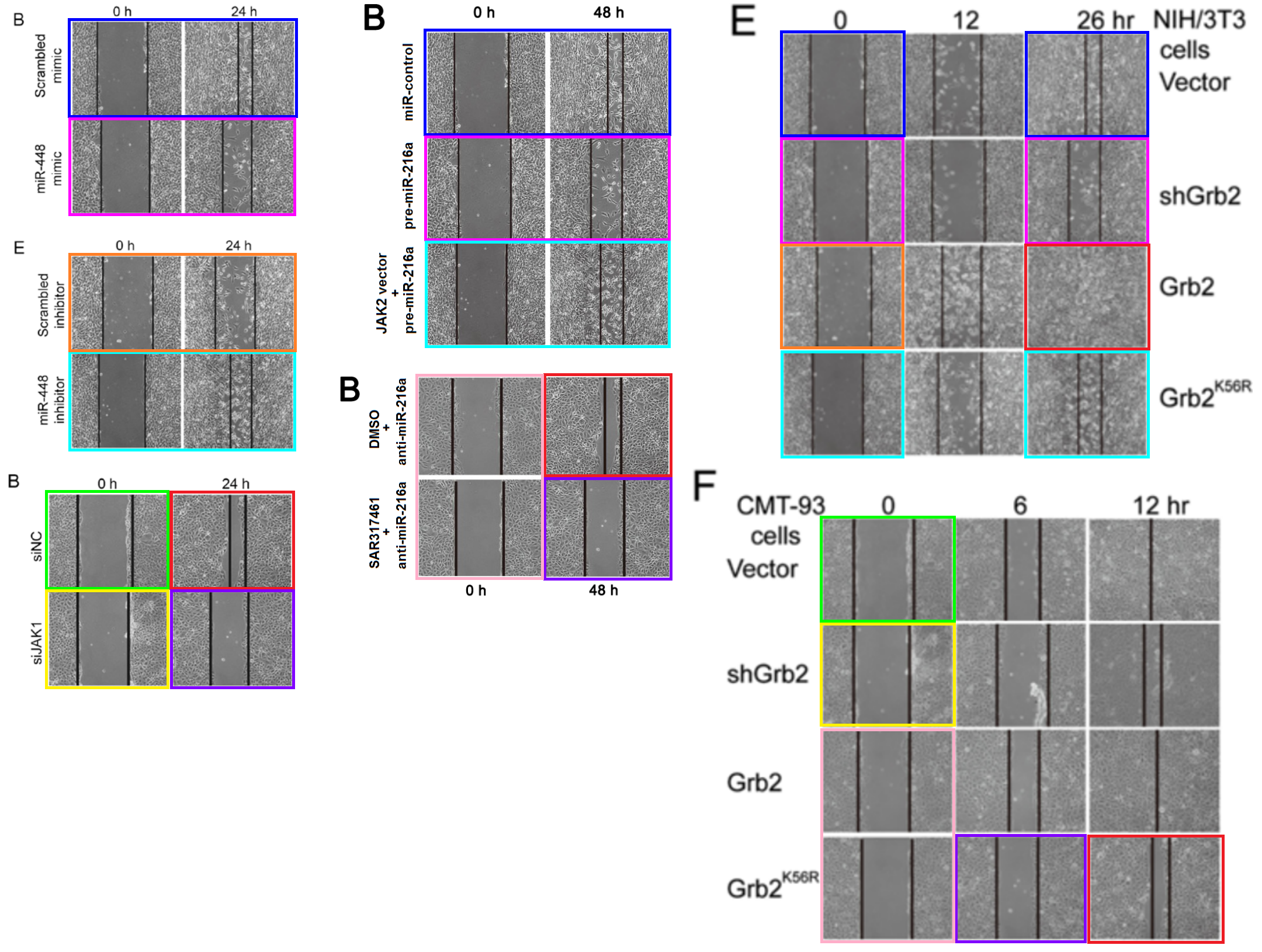
[middle] Figs 7B, 8B from “MicroRNA-216a inhibits the metastasis of gastric cancer cells by targeting JAK2/STAT3-mediated EMT process” (Tao et al 2017).
[right] Fig 4E/F from “SUMOylation of Grb2 enhances the ERK activity by increasing its binding with Sos1” (Qu et al 2014).
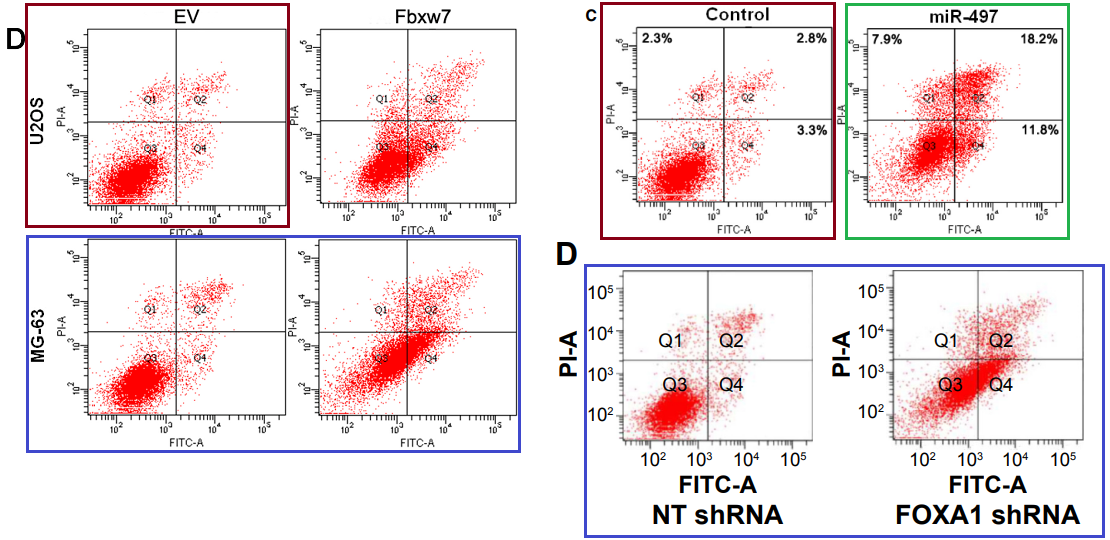
…And some flow-cytometry scatterplots.
[upper right] Fig 4 from “microRNA-497 inhibits cell proliferation and induces apoptosis by targeting YAP1 in human hepatocellular carcinoma” (Zhang et al, 2016) [retracted].
[lower right] Fig 4D from “Forkhead box protein A1 is a prognostic predictor and promotes tumor growth of gastric cancer” (Ren et al, 2015) [retracted].
Is there anyone from the Department of Hepatobiliary Surgery who doesn’t show up as coauthor of one of these paper-shaped malarkey vehicles? I have no idea how the faculty is structured, but one could speculate that “number of papers that one signs” is related somehow to “position in the departmental totem-pole”. As the dates on these papers progress from 2014 to 2021, some regular authors fade out while replacements arrive, but Kangsheng Tu is there from beginning to end.

Crucially for this context, the use of these bogus papers was not confined to the careers of the Hepatobiliary Surgery staff. A few were donated to colleagues at Departments of Emergency, and of General Surgery, but still within hospitals affiliated to Xi’an Jiaotong University. Others went further afield… to Zhejiang Provincial People’s Hospital; to Renji Hospital, School of Medicine, Shanghai Jiaotong University; to Fifth Affiliated Hospital of Guangzhou Medical University, Guangzhou… though as noted, some of these might be the work of plagiarists. The earliest evidence of shenanigans from this group is in Zheng et al (2012), in PLoS One: an international collaboration with the main author from the Mayo Clinic.
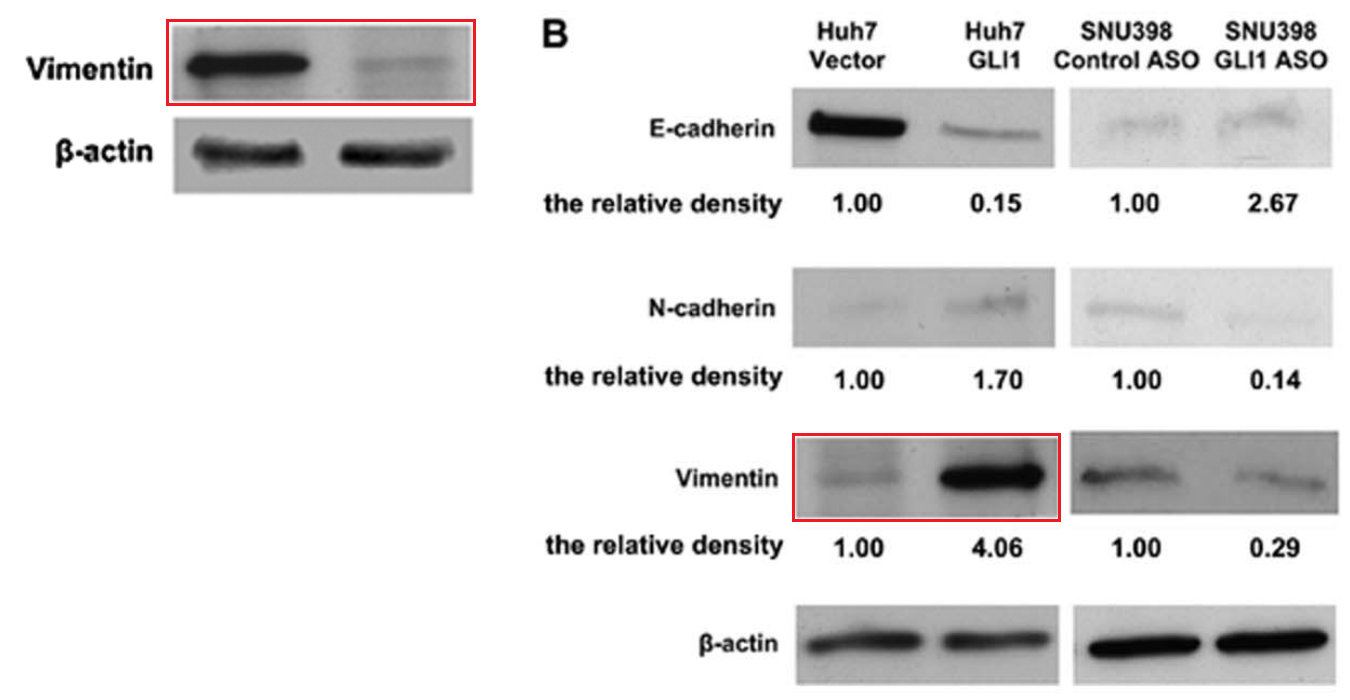
[right] Fig 4B from “The Transcription Factor GLI1 Mediates TGFb1 Driven EMT in Hepatocellular Carcinoma via a SNAI1-Dependent Mechanism” (Zheng et al 2012)..
The editors of FEBS Open Bio engaged with some of the PubPeer critiques, reporting in July 2022 that “The journal is aware of these issues and is currently investigating“, though they don’t seem to have brought their investigations to the attention of their wider readership by issuing Expressions of Concern.
Again, authors have been responsive… Errors were made, and goats were scaped, and if “Throwing to the Wolves” ever becomes an Olympic event, then Xi’an Jiaotong University could field a world-beating team. There is a general sense of intense behind-the-scenes lobbying among the various customers, choosing which papers to sacrifice in the hope of salvaging others. Zhikui Liu blamed colleagues for the falsifications in three cases, then blamed himself twice thrice, then took the rap for everything.

The Fragments of papermills
Going back to retractions: The ‘bowtie’ style of algorithm-generated WB facsimiles is increasingly encountering editorial side-eye and suspicion. That is, the style is on the path to perdition, following the now-deprecated Ransom-Note, string-of-sausages and ‘blobby’ conventions.
The scientific sea of miR- and exosome-related knowledge
“Whole cohorts of peer-reviewers have been trained to view all these mannerist stylings as what western blots should look like. […] It will be a challenge to convince them otherwise.” – Smut Clyde.
It is a safe bet that in their current work, the ‘Contractor’ millers have adopted a new generative style, not yet recognised and delineated. I had planned to write about another corpus consisting of some possible examples (though these might just be the work of another cottage) – but this post is long enough already.

For the same reason of “excess length”, several other corpora of artisanal fakes have also wound up on the cutting-room floor. At the moment, 12 small networks of interlinked fabrications are collated in a “Fragments” spreadsheet. Some of those might become spreadsheets in their own right, if they grow large enough, and if I can summon up the energy. This is all in a state of flux, and some of these corpora might conflate the work of more than one studio; conversely, what are now separate fragments within ‘Fragments’ might merge.
I claimed at the start that simple hallmarks are harder to find for the larger production lines. This was of course a lie deliberate mistake to see who was paying attention.
The ‘Problematic Paper Screener‘ (Cabanac et al 2022) has a Feet-Of-Clay operating mode that scans the References sections of papers for known papermill products. Of course uncritical authors might cite one or two forgeries when the Abstracts seem to fit their Just-So story or the theme of their literature review, but artisanal papermillers are even less critical (and better acquainted with the penumbral literature of ‘parascience’), so the referrals from ‘Feet-Of-Clay’ turn out to be dominated by their products… at least in the domains of biotech.
Perhaps this is not so much an indication that (some) papermills just grab References at random, and more an indictment of the prevalence of egregious forgeries in the literature, so that several are swept up into each of those random Reference-section assortments.
Retractions, and the “Transwell” papermill
Overnight, FEBS Open Bio progressed from “awareness of the concerns” and “currently investigating” nine papers, to retracting them on 15 November 2022.
Three dealt with fabrications from the Xi’an Jiaotong University cluster. Authors from that group seized on the retractions to defend a subsequent paper in they recycled images.
“These projects were going on at about the same time. The project (Lymphocyte-specific protein 1 inhibits the growth of hepatocellular carcinoma by suppressing ERK1/2 phosphorylation) unintentionally included an image from the project (Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/β-catenin signal pathway in hepatocellular carcinoma) when storing and organizing data due to the similarity of appearance and file name. “
One retracted a product of the “Colony retraction” cottage papermill.

Now the out-takes from this post included what was described in a PubPeer thread as “Nothing to see here… just a growing network of papers from unrelated teams of authors, illustrated with transwell invasion / migration images drawn from a single pool.” Inspiration failed when it came to naming this cottage so the inevitable spreadsheet is just called “Transwell”. Only 14 entries so far… two of its products found their way into EXCLI Journal, so if I were that journal’s editor, I would check the back-catalog to see how many others slipped under the radar. Customers are distributed across China with no obvious focus that might point to the source. Anyway, off the cutting-room floor it goes, and back into the post!
Imagination continued to fail, which is why another corpus is just Section #5 in the ‘Fragments’ spreadsheet, currently in rows 43-68 (I remind readers that naming rights are available). Three of that studio’s products were also retracted.

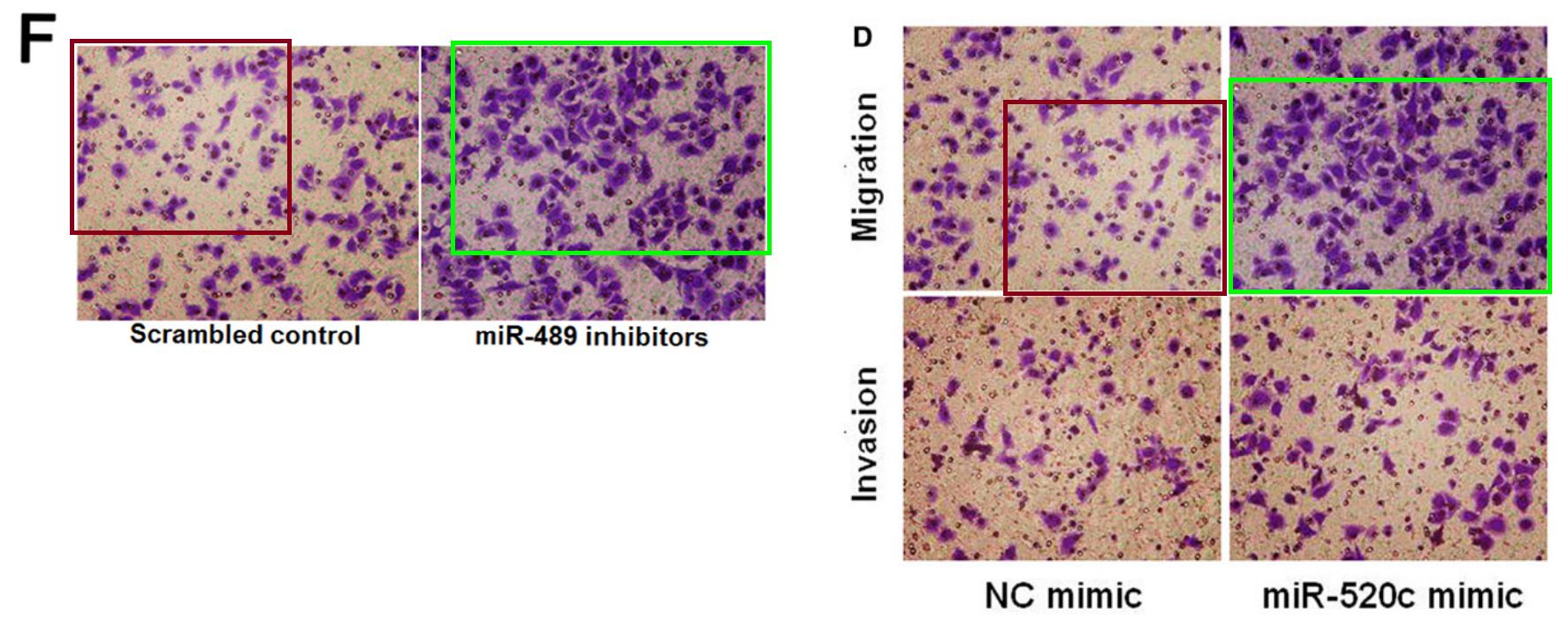
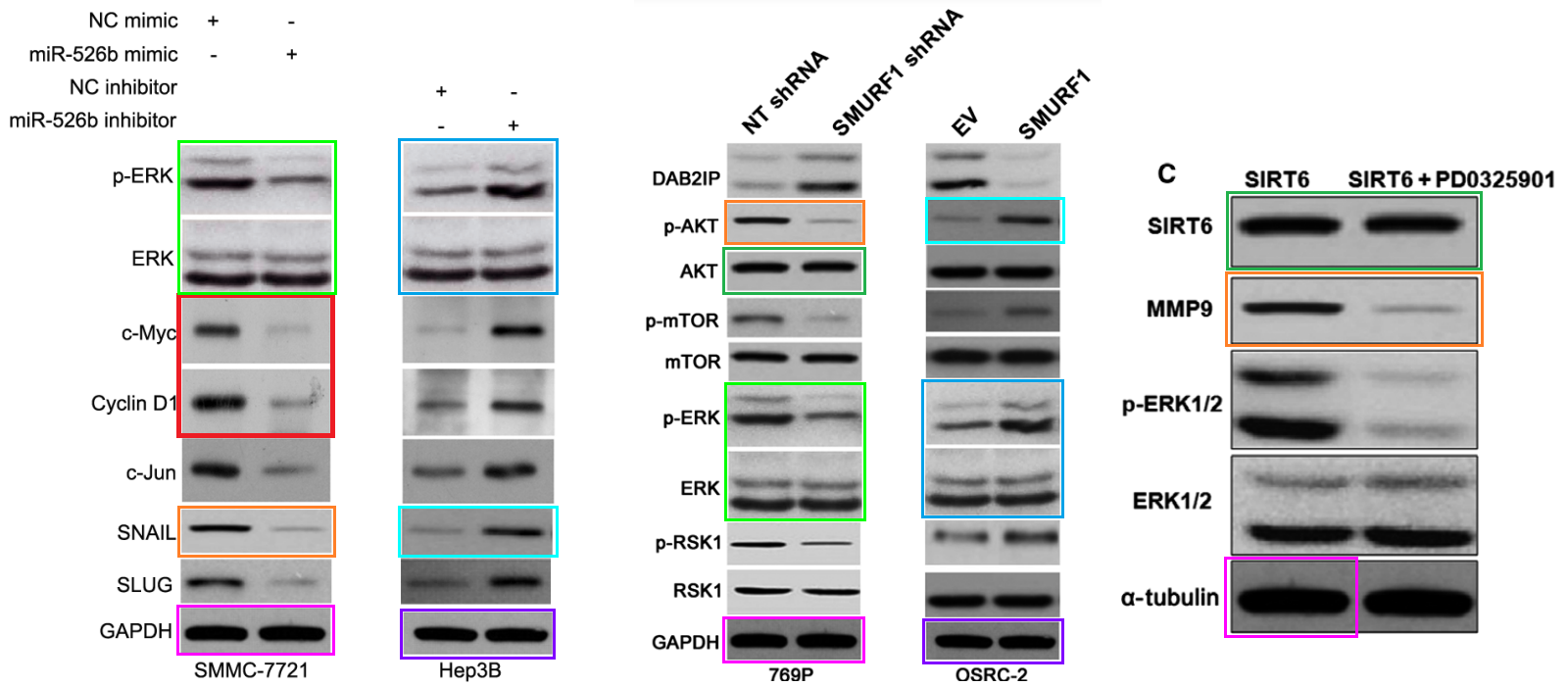
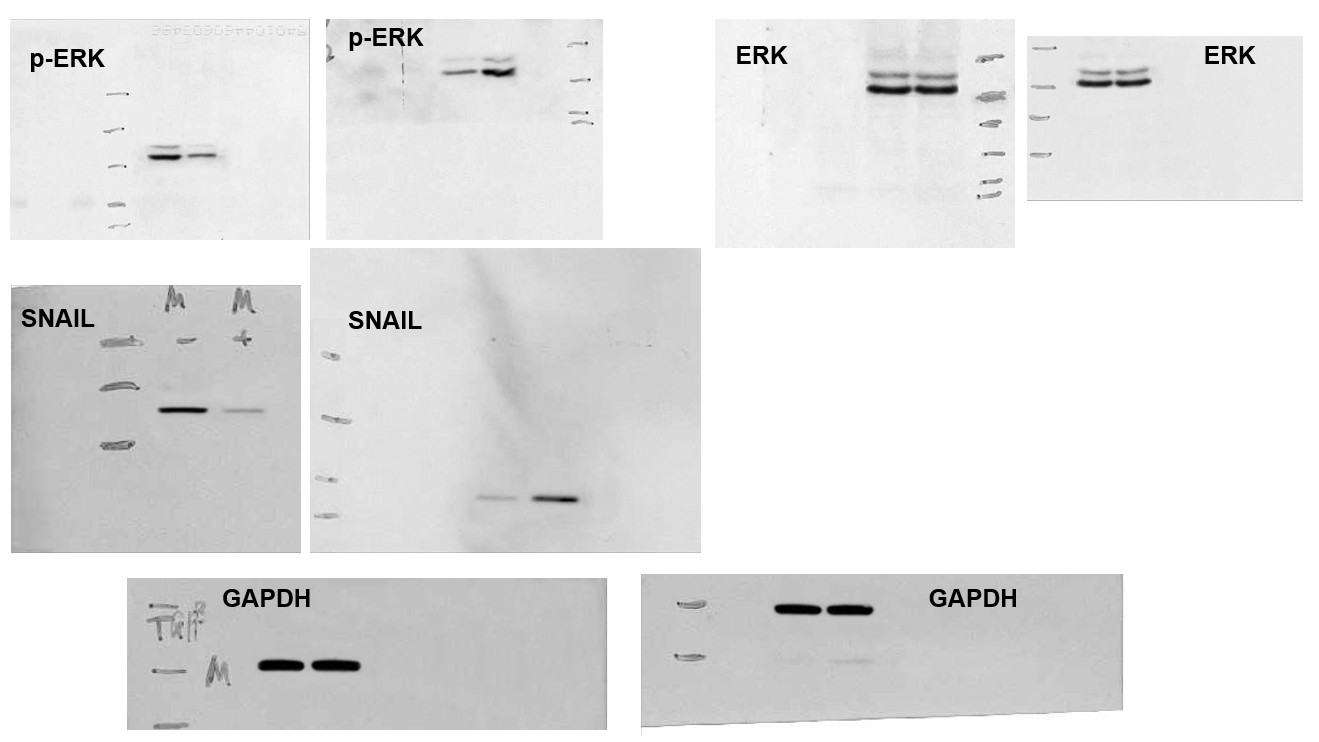
[middle] Fig 5 from “Ubiquitin ligase SMURF1 functions as a prognostic marker and promotes growth and metastasis of clear cell renal cell carcinoma” (Ke et al, 2017) [retracted].
[right] Fig 6C from “Sirtuin 6 contributes to migration and invasion of osteosarcoma cells via the ERK1/2/MMP9 pathway” (Lin et al, 2017) [retracted].
Within this #5 corpus we find “microRNA-526b serves as a prognostic factor and exhibits tumor suppressive property by targeting Sirtuin 7 in hepatocellular carcinoma” (Liu et al, 2017). This encountered criticism in PubPeer, on account of several Figures having featured in earlier products of the studio. Corresponding Author Qiuran Xu came to the paper’s defence, providing evidence of possession of originals, while arguing that the research collaboration had been underway since 2012 but unspecified miscreants had pilfered copies of the images along the way:
“Our project started in 2012, the experiments didn’t go well, the project changed hands several times, and it was finally published on Oncotarget in 2017“.
I do not want to get into the plausibility of this explanation. I will only note that images are occasionally exchanged between the #5 studio and the “Shaanxi” studio at Xi’an Jiaotong University. This directs our attention to the other Corresponding Author on the paper, Yingmin Yao, who is one of the Xi’an Jiaotong University circle.

Donate to Smut Clyde
If you liked Smut Clyde’s work, you can leave here a small tip of 10 NZD (USD 7). Or several of small tips, just increase the amount as you like (2x=NZD 20; 5x=NZD 50). Your donation will go straight to Smut Clyde’s beer fund.
NZ$10.00
















Thank you to all the sleuths doing the hard work to uncover these schemes.
LikeLike
They know who they are.
LikeLike
I know: Prof Jennifer Byrne and Jana Christopher. And Ivan Oransky, and journalists of Science and Nature. And everyone else, but not you losers.
LikeLike
Great work!!
LikeLike