On the Iberian peninsula, there seems to be a tradition to give well-connected scientists suspected (or even convinced) of data fudging an award. In Spain, Carlos López-Otín, Professor of Biochemistry and Molecular Biology at the University of Oviedo, was given a Mentoring Award from the elite journal Nature, on recommendation from Spanish academia and despite evidence of data irregularities in his papers. This prompted my readers, in particular the famous pseudonymous data integrity sleuth Clare Francis, to comment on on PubPeer and on my site (as “Zebedee”) with additional evidence, which made Lopez-Otin’s scientific credibility look progressively worse and worse, with each new post.
Eventually, an image of a Northern blot (showing expression of mRNAs which code for proteins) was found to appear recurrently across several papers from that Oviedo lab, where the authors pretended it was a newly produced analysis. In reality, it was a “library” loading control reused so the authors could re-run same RNA gel of human tissue lysates over the years and never check ever again what they have actually loaded on their gels. Eventually Lopez-Otin et al even stopped caring what order of samples that original loading control had.
Clare Francis was soon joined on his quest for the Perennial Northern Blot of Oviedo by Elisabeth Bik, famous microbiology blogger and image duplication detective, and my regular contributor (also pseudonymous) Smut Clyde, who now presents you the findings of no less than 23 appearances of that same northern blot in 23 publications from Lopez-Otin’s lab in the guest post below. It is just as convincing as if the Spanish actor Antonio Banderas appeared in 23 different films still dressed in same costume from his 1995 hit Desperado, carrying same guitar case. Incidentally, also Lopez-Otin’s Perennial Northern Blot made its first appearance at around that year.

The cancer researcher Lopez-Otin is an actual real-life celebrity in Spain. On 29 June 2017, he helped the King of Spain open the Princess of Girona Foundation Awards ceremony, together with another Spanish celebrity of the creative art of the pretend, Antonio Banderas. The Princess Foundation wrote about their 2017 gala host:
“Carlos Lopez-Otín is an academician of the European Academy and the Royal Academy of Sciences of Spain, and Doctor Honoris Causa by several Spanish and foreign universities. Throughout his scientific career he has received several awards such as the European FEBS Prize for Biochemistry, the DuPont Award for Life Sciences, the “Carmen and Severo Ochoa” Award, the Mexico Prize for Science and Technology, King Jaime I Prize Research and the National Research Award “Santiago Ramón y Cajal”
That multiple award winning research star is also EMBO member, just like some other of his Spain-based colleagues of questionable research integrity are: Maria Pia Cosma and Pura Munoz-Canoves. These two also regularly receive juicy research grants and recognition: Munoz-Canoves’ most recent was the “Vanguardia de la Ciencia” award, while Cosma was given in 2016 “Ciutat de Barcelona” award. Another Spanish researcher with shady data in his papers is Manel Esteller, he also gets awarded regularly, in 2016 it was the Catalonia International Prize from (now fugitive) President Puigdemont. In Portugal, Esteller’s former PhD student and now zombie scientist Sonia Melo was given a Prémio FAZ Ciência award from Fundação AstraZeneca, only two months ago. There are surely more of such questionable Iberic awardees, readers are welcome to salute these stars of Photoshop in the comment section.
I personally have a theory that in this way the system of Iberian academia announces who is untouchable, in order to intimidate critics of research misconduct in their own ranks. Probably a pathetic left-over from the fascist past of Spain and Portugal. The subliminal message is: yes, we all know what these award-winners did to create those big papers and we don’t mind at all. The award-giving farce shows who the role models are and what Spanish and Portuguese scientists are expected to do with their silly notions of research integrity: shove it, start making big papers whatever it takes, or lose your job. In fact, not even the arch-zombie scientist Susana Gonzalez had to suffer unemployment, unlike masses of honest young Spanish scientists.
A Perennial Northern Blot, by Smut Clyde
The title of this post refers to the famously picaresque Western blot belonging to a Brazilian diabetes researcher. 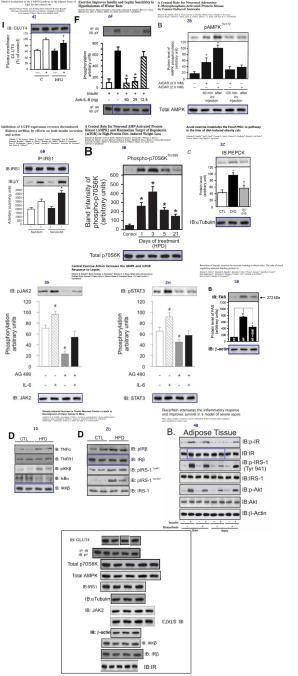 In its protean versatility, Mario Saad‘s pentadecaplicating blot could transform itself into any protein — tubulin, actin, GLUT4, IRS1 — from any combination of source conditions. It thereby appeared in at least 15 versions, spread across 10 papers in “an intricate publishing web“, serving as the loading control in that many different experiments (that is, as a measure of the total level of extracted protein, for normalising the measurements of the protein of interest). In my imagination it spoke with the voice of Robin Williams. This site forwarded a report on the Wandering Western… the ensuing saga included editorial Expressions of Concern, lawsuits, an investigation by Saad’s university that saw no evidence of misconduct, and 13 retractions so far (RetractionWatch are keeping score).
In its protean versatility, Mario Saad‘s pentadecaplicating blot could transform itself into any protein — tubulin, actin, GLUT4, IRS1 — from any combination of source conditions. It thereby appeared in at least 15 versions, spread across 10 papers in “an intricate publishing web“, serving as the loading control in that many different experiments (that is, as a measure of the total level of extracted protein, for normalising the measurements of the protein of interest). In my imagination it spoke with the voice of Robin Williams. This site forwarded a report on the Wandering Western… the ensuing saga included editorial Expressions of Concern, lawsuits, an investigation by Saad’s university that saw no evidence of misconduct, and 13 retractions so far (RetractionWatch are keeping score).
The present case also concerns re-use of a loading control, but this time featuring a Northern blot. The compass-point tradition for naming gel-electrophoresis techniques began with Sir Edwin Southern, pioneer of Southern blotting, for this is how humor works in molecular biology. It has been explained to me that Northern blots do not directly measure the popularity of a protein in the cellular economy; instead, mRNA (encoding for a protein) is the chemical species, extracted from various sources (lanes), and spread out into bands according to molecular weight. Then transferred (blotted) from the electrophoresis gel to a filter for stability, and detected by inducing the mRNA to bind to a matching and radiotracing DNA probe.
So in this case, a team of researchers have a bank of 28 “cell smoothies”: two sets of eight tissue types, one set of eight cancer-cell lines, and four fetal-tissue samples. In a series of papers published over a decade, the team have characterised numerous proteins from within the self-organising complexity of the human cell — sequencing the DNA for each protein and specifying its chromosomal location, describing its role within that complexity, and checking which tissues express it (which depends on which genes remained active in each lineage of cells that differentiated and specialised and became a tissue). That is to say, the Northern Blots were just one aspect of the papers, and they are all outside my comfort grade and above my pay zone.

Each study took a few drops from the stored samples, blotted it (“Filters containing about 2 µg of polyadenylated RNAs from the indicated human tissues”), and probed for the mRNA of choice. But there are limits to the precision that a pipette can provide — even in the hands of a trained gene-modified laboratory monkey — so the final stage is to wash the probe DNA out of the filter and probe it again for Actin (a background “housekeeping” protein, required by cells to maintain their architecture, unless they are dead) to correct for the actual aliquots that were used. Thus papers in this sequence typically include a phrase along these lines:
“Filters were subsequently hybridized with a human actin probe to ascertain differences in RNA loading”.
It is conceivable, however, that this phrase was repeated from the first paper, along with the loading blot itself. Comparing 23 papers, there appears to be one original blot for each bank: four blots, which are variously compressed and clipped according to the exigencies of publication, and varying also in exposure, rather than a separate measurement after each separate exercise in tissue localisation. The sources are ‘Zebedee’, commenting on threads at this site; anonymous contributors to PubPeer threads; Elisabeth Bik; and myself.
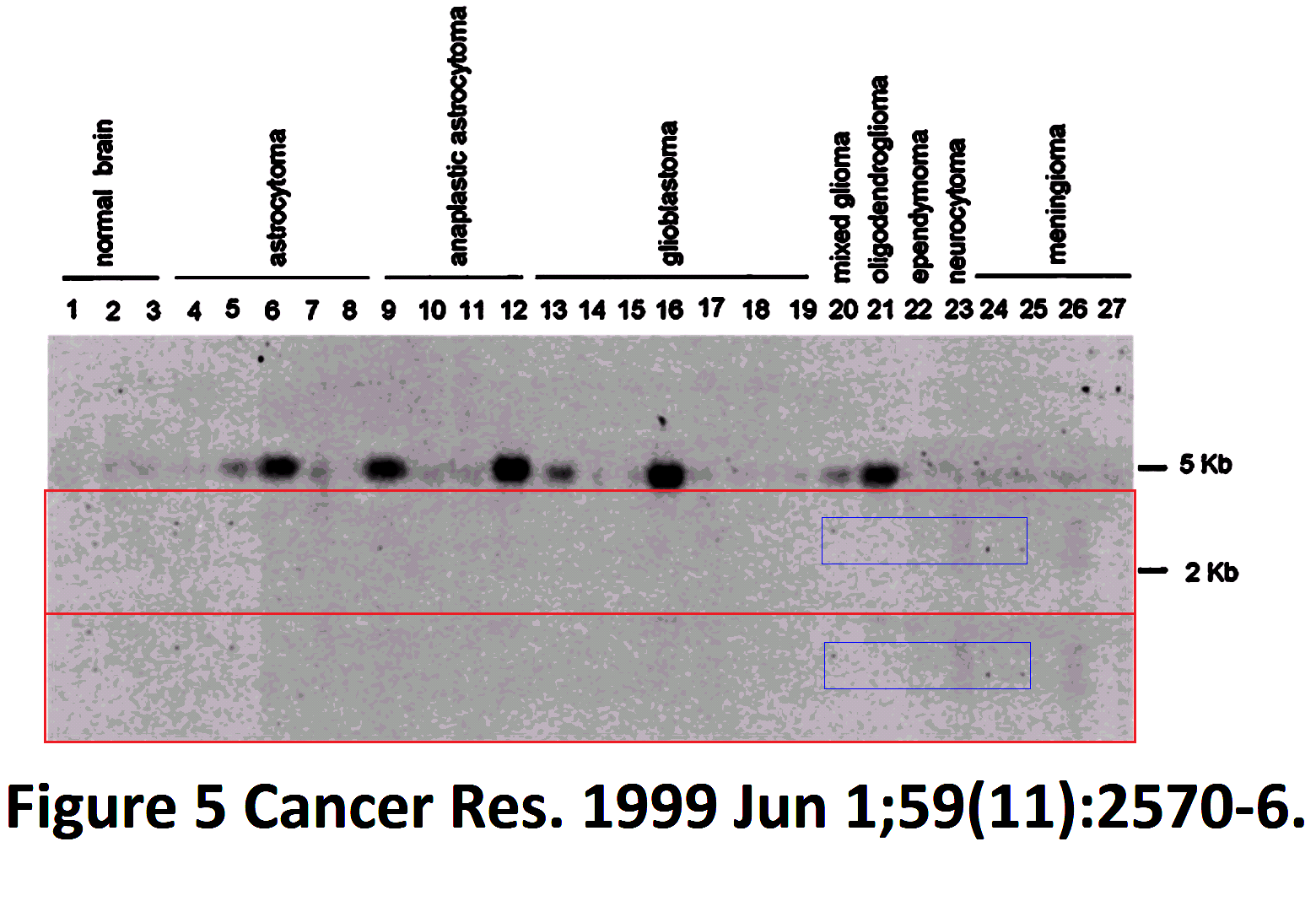
This comes to our notice because a 23-fold replication beats the 15-fold record of the Brazilian wanderer. Crucially, though the possible copies are consistently identified as Actin, and the authors have tried to label the sources of the lanes consistently. The reuse of a ‘loading library’ is deprecated, but this does not begin to approach the problematic level of the Brazilian Western: there was no attempt to mislead (other than the claim that the control in each study was specific to it, made subsequently to the data to be controlled). It is a perennial blot, always in the same place, rather than a wanderer or vagrant.

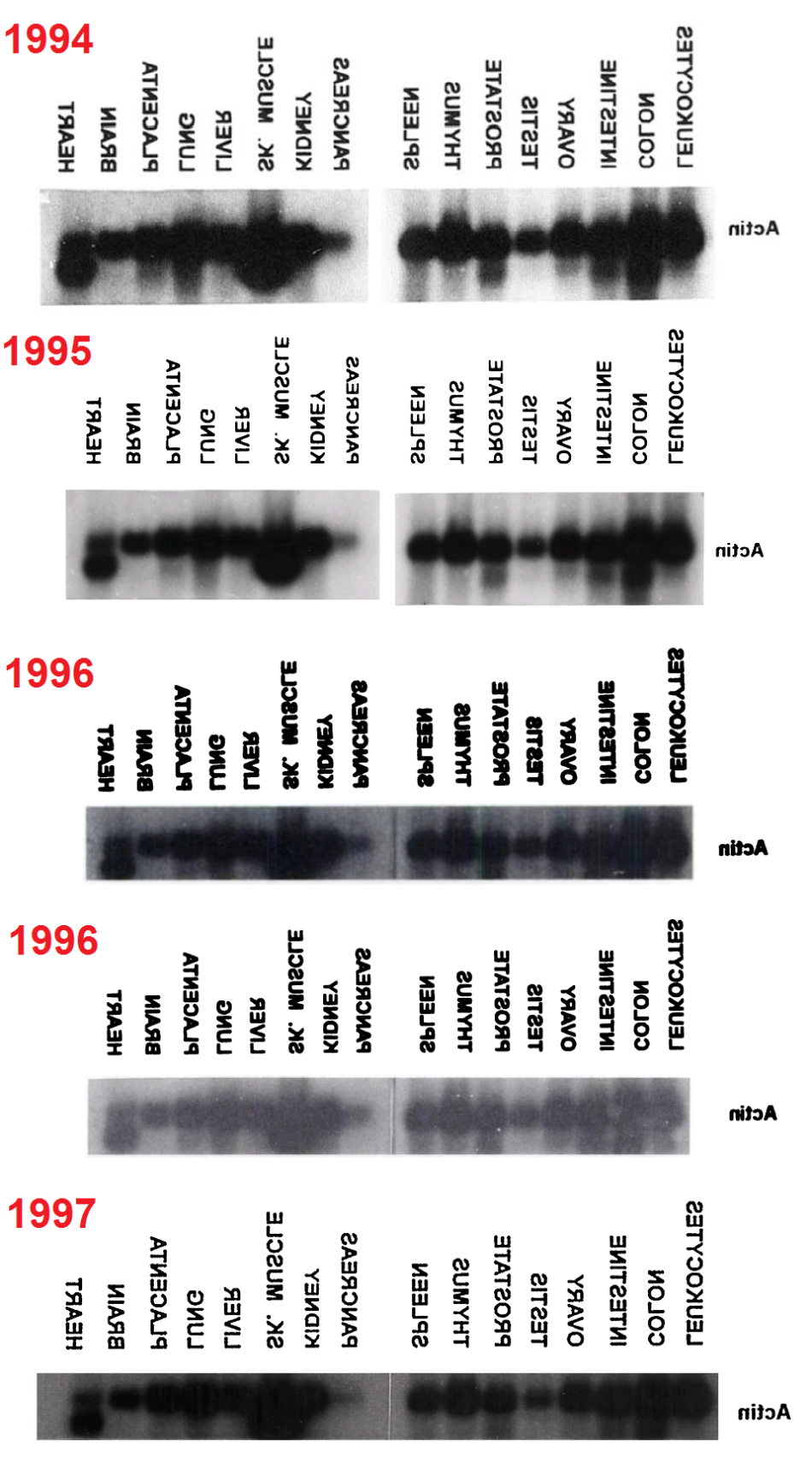
Regrettably, the labelling of lanes was not as consistent as was intended. In a 2003 appearance of the fetal-tissue blot, it was flipped horizontally relative to the lane labels, as marked with a red box in the Figures. Note that in some publications the lanes are listed in reverse order — from Leucocytes to Heart rather than vice versa — and in the Figures I have flipped each band and labels in such cases, to keep a single sequence of tissue types (hence the mirror-image text in places).
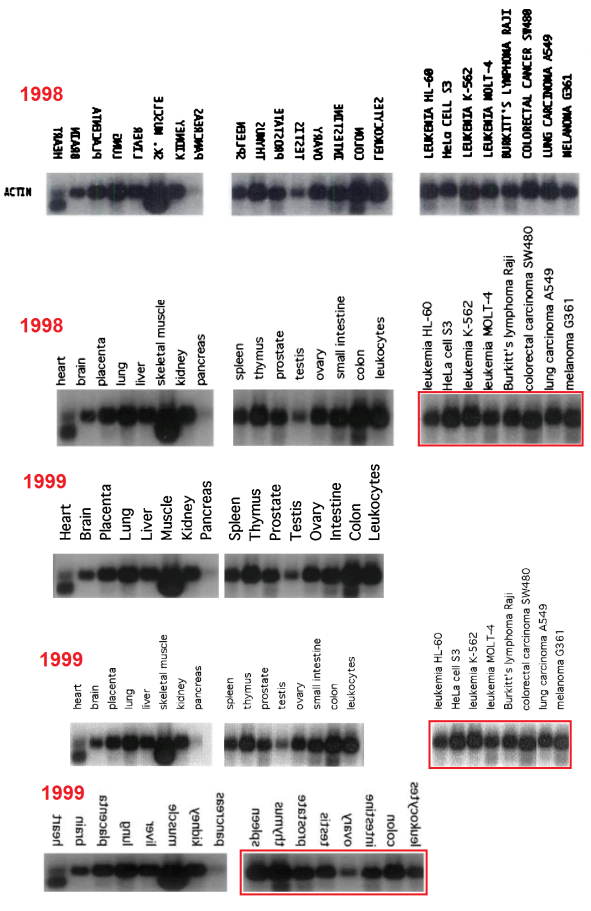
Red boxes were also necessary in some cases where the cancer-cell blot was flipped relative to its lane labels, and for the #2 array of tissue cells in the 1999 paper. In addition, that blot was rotated through 180° from 2001 onwards (so that the Actin background for Thymus cells becomes that of Colon cells, and vice versa, while Testes and Ovary change places, and Spleen with Leucocytes). This is marked with orange boxes. One can only hope that these pictorial labelling issues did not extend into the measurements of Actin from the blots, as used in the quantitative results.
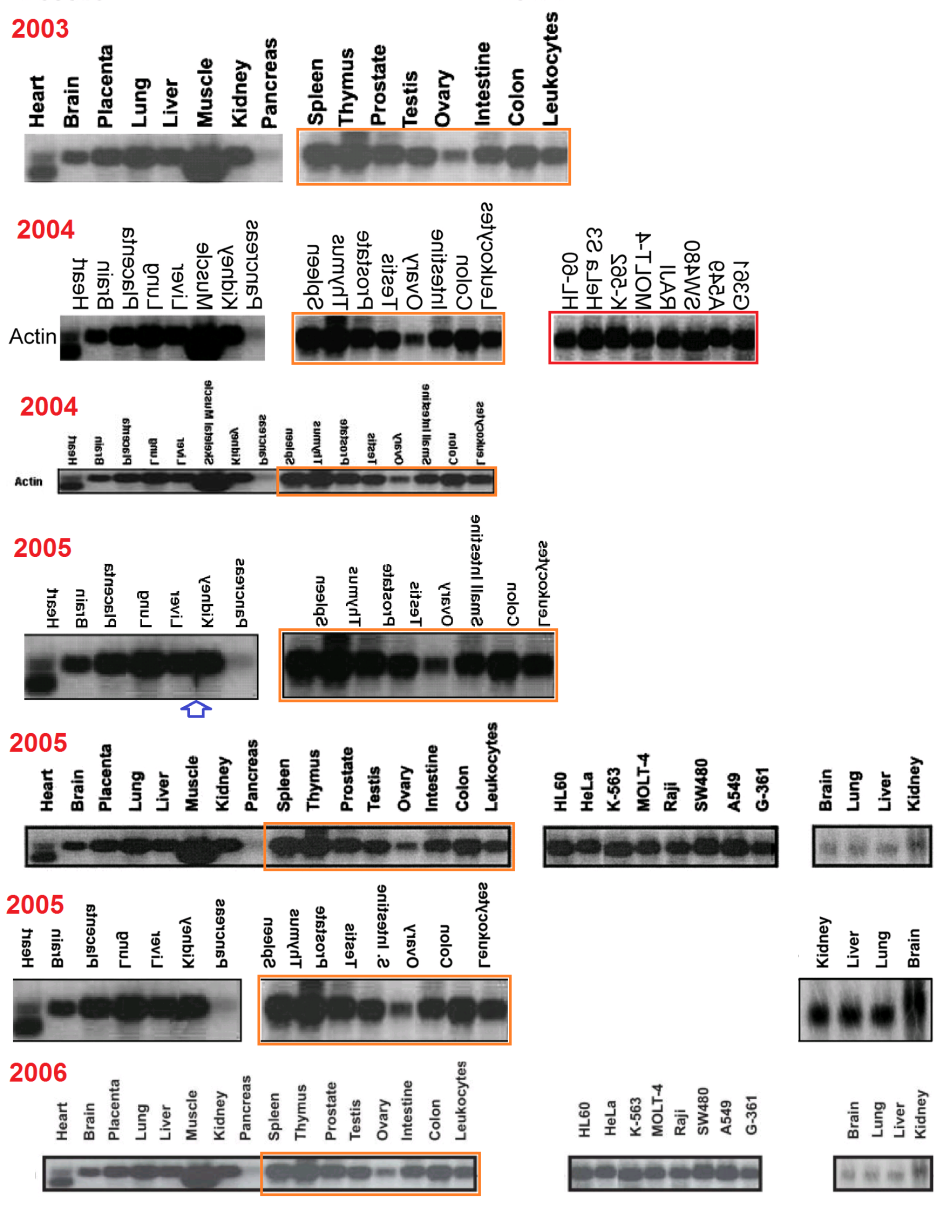
Finally, two blue arrows mark the omission of ‘Pancreas’ and ‘Skeletal muscle’ from one study each, with the loading band spliced to remove that lane.
I am going to play ‘good cop’ here, and propose that the corner-cutting absence of study-specific controls probably made little difference to the results. Corrigenda to the paper acknowledging the use of archival controls would be appropriate (along with correction of any flipped and rotated bands). Other issues have been raised about other figures in some of the papers, but I do not address those here.
Details of the 23 publications follow. We are still hopeful of finding a few more examples of the Perennial Northern Blot.
- 1994. “Human cathepsin O. Molecular cloning from a breast carcinoma, production of the active enzyme in Escherichia coli, and expression analysis in human tissues“, Velasco et al; J Biol Chem., 269(43):27136-42.
- 1995. “Cloning and expression analysis of a novel human serine hydrolase with sequence similarity to prokaryotic enzymes involved in the degradation of aromatic compounds“, Puente & López-Otín; Journal of Biological Chemistry 270, 12926-12932. DOI 10.1074/jbc.270.21.12926 Figure 5.
- 1996. “Cloning and Expression Analysis of Human Bleomycin Hydrolase, a Cysteine Proteinase Involved in Chemotherapy Resistance“, Ferrando et al.; Cancer Research 56: 1746-1750. PMID: 8620487
- 1996. “Molecular Cloning of a Novel Membrane-type Matrix Metalloproteinase from a Human Breast Carcinoma“, Puente et al; Cancer Research 56:944-949.
- 1997. “Identification and characterization of a novel human matrix metalloproteinase with unique structural characteristics, chromosomal location, and tissue distribution“, Pendás et al; J Biol Chem. 272(7):4281-6. doi: 10.1074/jbc.272.7.4281 Figure 7.
- 1998. “Cathepsin L2, a Novel Human Cysteine Proteinase Produced by Breast and Colorectal Carcinomas“, Santamaría et al; Cancer Res. 58(8):1624-30.
- 1998. “Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location“, Santamaría et al; J Biol Chem. 273(27):16816-23. doi: 10.1074/jbc.273.27.16816 Figure 5.
- 1999. “Cloning and characterization of human MMP-23, a new matrix metalloproteinase predominantly expressed in reproductive tissues and lacking conserved domains in other family members“, Velasco et al; J Biol Chem. 274(8):4570-6. doi: 10.1074/jbc.274.8.4570 Figure 6.
- 1999. “Molecular cloning and structural and functional characterization of human cathepsin F, a new cysteine proteinase of the papain family with a long propeptide domain“, Santamaría et al; J Biol Chem. 274(20):13800-9. doi: 10.1074/jbc.274.20.13800 Figure 6.
- 1999. “Identification and Chromosomal Location of Two Human Genes Encoding Enzymes Potentially Involved in Proteolytic Maturation of Farnesylated Proteins“, Freije et al; Genomics 58, 270–280. DOI: 10.1006/geno.1999.5834
- 2000. “Human MT6-matrix metalloproteinase: identification, progelatinase A activation, and expression in brain tumors“, Velasco et al; Cancer Research 60, 877–882. pubmed: 10706098
- 2001. “Identification, Characterization, and Intracellular Processing of ADAM-TS12, a Novel Human Disintegrin with a Complex Structural Organization Involving Multiple Thrombospondin-1 Repeats“, Cal et al; Journal of Biological Chemistry 276, 17932-17940. doi: 10.1074/jbc.M100534200 Figure 5.
- 2002. “Matriptase-2, a Membrane-bound Mosaic Serine Proteinase Predominantly Expressed in Human Liver and Showing Degrading Activity against Extracellular Matrix Proteins“, Velasco et al; J Biol Chem. 277(40):37637-46. doi: 10.1074/jbc.M203007200 Figure 8.
- 2002 “Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains“, Cal et al; Gene 283 49-62. doi: 10.1016/S0378-1119(01)00861-7
- 2003. “Polyserase-I, a human polyprotease with the ability to generate independent serine protease domains from a single translation product“, Cal et al; PNAS 100(16): 9185–9190. doi: 10.1073/pnas.1633392100 Figure 4.
- 2003. “Human Autophagins, a Family of Cysteine Proteinases Potentially Implicated in Cell Degradation by Autophagy“, Mariño et al; Journal of Biological Chemistry 278, 3671-3678. doi: 10.1074/jbc.M208247200 Figure 3.
- 2003. “Identification and Characterization of ADAMTS-20 Defines a Novel Subfamily of Metalloproteinases-Disintegrins with Multiple Thrombospondin-1 Repeats and a Unique GON Domain“, Llamazares et al; Journal of Biological Chemistry 278(15):13382-13389. doi: 10.1074/jbc.M211900200 Figure 4.
- 2004. “Identification and Characterization of Human and Mouse Ovastacin“, Quesada et al; JBC 279 (25) 26627-26634. doi: 10.1074/jbc.M401588200 Figure 3.
- 2004. “Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases“, Queseda et al; Biochemical and Biophysical Research Communications 314, 54-62. doi: 10.1016/j.bbrc.2003.12.050
- 2005. “Identification of Human Aminopeptidase O, a Novel Metalloprotease with Structural Similarity to Aminopeptidase B and Leukotriene A4 Hydrolase“, Díaz-Perales et al; Journal of Biological Chemistry 280, 14310-14317. doi: 10.1074/jbc.M413222200 Figure 4.
- 2005. “Human Polyserase-2, a Novel Enzyme with Three Tandem Serine Protease Domains in a Single Polypeptide Chain“, Cal et al; JBC 280, 1953-1961. doi: 10.1074/jbc.M409139200 Figure 3.
- 2005. “Identification and Characterization of Human Archaemetzincin-1 and -2, Two Novel Members of a Family of Metalloproteases Widely Distributed in Archaea“, Diaz-Perales et al; JBC 280(34):30367-30375. doi: 10.1074/jbc.M504533200 Figure 4.
- 2006. “Identification and characterization of human polyserase-3, a novel protein with tandem serine-protease domains in the same polypeptide chain“, Cal et al; BMC Biochemistry. doi: 10.1186/1471-2091-7-9 Figure 7.
Update 7.05.2018. Based on reader comment:


Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism will be most appreciated!
€5.00


Pingback: Lopez-Otin and Kroemer: birds of a feather flock together – For Better Science
Pingback: Lopez-Otin and Daley retract Nature Cell Biology paper – For Better Science
Pingback: Spanish elites rally in support of data manipulation – For Better Science
Pingback: Frederique Vidal, Minister for Research and gel band duplication – For Better Science
Pingback: El caso López-Otín de fraude científico - La Ciencia de la Mula Francis
Pingback: When gel bands go marching in, by Elisabeth Bik – For Better Science
Pingback: Carlos Lopez-Otin and the revoked Nature Mentoring Award – For Better Science