There is only one Smut Clyde, but the papermill fraudsters he chases are multitude! You will see it is not just the figures which get recycled among papers from totally unrelated groups of Chinese “authors”. A pro-tip to journal editors: do check if the corresponding author’s email address makes any sense at all.
In last week’s Friday Shorts, you may have read of Smut Clyde’s and Tiger BB8’s discovery of fake Georgians, i,e, Chinese papermill fraudsters who created a fictional email account for a university in Tbilisi, and even made-up identities of fictional local coauthors.
Now Smut Clyde continues with an investigation of Chinese Doppelgängers who share email addresses and of course also figures across research fields, time and space. Here is the necessary Sheets file for you, with over 220 papermilled entries.
And now, Smut Clyde will past bilocation and Doppelgängers.

Mirrors and copulation are abominable because they increase the number of mice
By Smut Clyde
It is never a pleasant experience to encounter two papers that seek to enhance the credibility of their separate narratives (otherwise bald and unconvincing) by showing the same identity parades of excised tumours, and the same murine crime scenes of donors of those tumours. Like discovering that what one thought was a window is actually a mirror, or maybe vice versa.
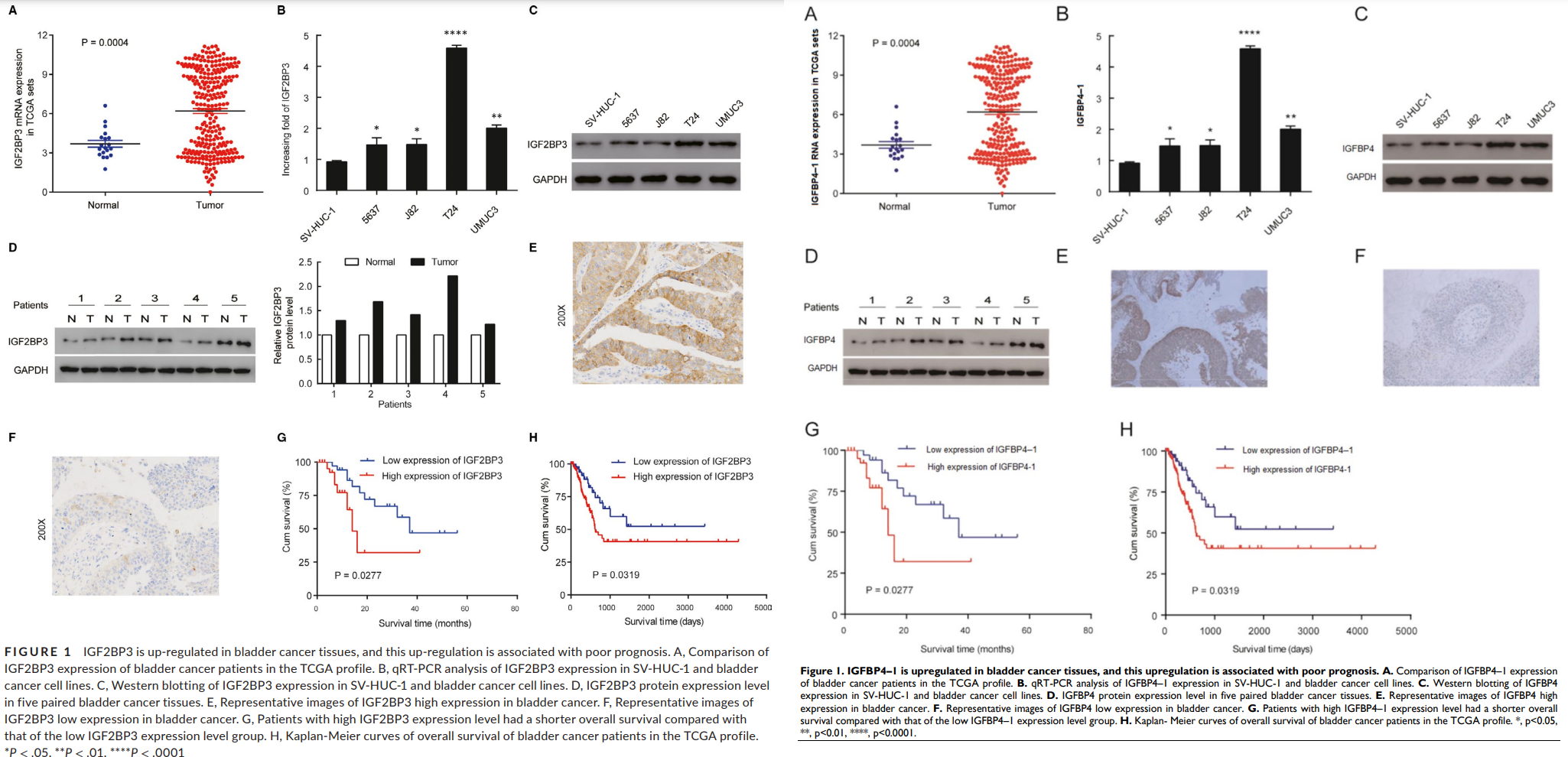
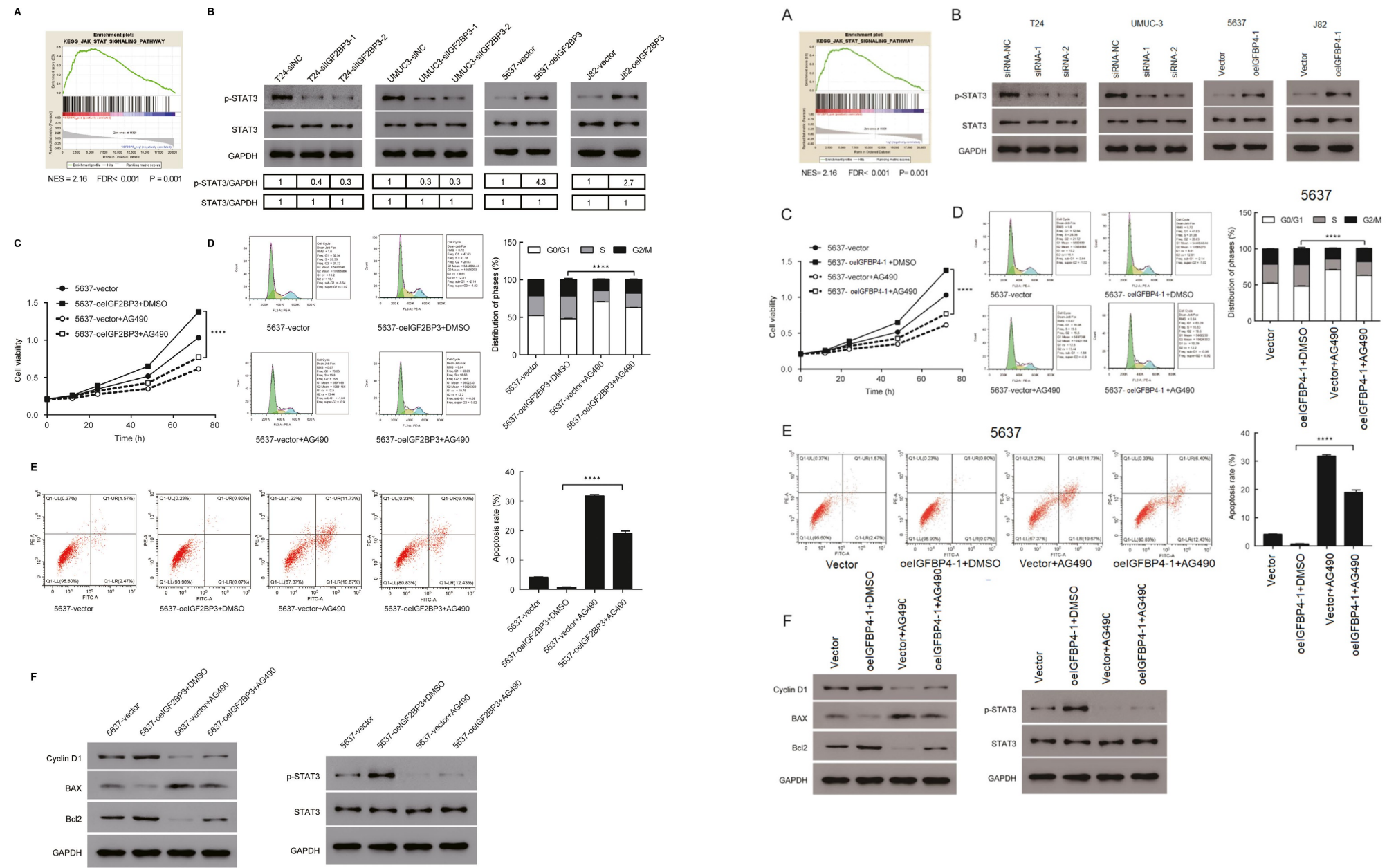
[right] Fig 1 from “LncRNA IGFBP4-1 promotes tumor development by activating Janus kinase-signal transducer and activator of transcription pathway in bladder urothelial carcinoma” (Li et al 2020).
To say nothing of “CRISPR-Cas13a Targeting the Enhancer RNA-SMAD7e Inhibits Bladder Cancer Development Both in vitro and in vivo” (Che et al 2020), containing third copies of those images:
 Li et al and Che et al have declined invitations to comment on these coincidences in the corresponding Pubpeer threads. On behalf of Huang et al., Gongxian Wang brought the receipts to claim precedence, establishing that in January 2020 they had shopped their version to Frontiers in Oncology before switching to Journal of Cellular & Molecular Medicine as its Forever Home. So one possibility is that the Frontiers reviewers retained pictorial material from the manuscript for their own version. There are other explanations, though, and they have the advantage that the process of explaining them will take us past bilocation and Doppelgängers.
Li et al and Che et al have declined invitations to comment on these coincidences in the corresponding Pubpeer threads. On behalf of Huang et al., Gongxian Wang brought the receipts to claim precedence, establishing that in January 2020 they had shopped their version to Frontiers in Oncology before switching to Journal of Cellular & Molecular Medicine as its Forever Home. So one possibility is that the Frontiers reviewers retained pictorial material from the manuscript for their own version. There are other explanations, though, and they have the advantage that the process of explaining them will take us past bilocation and Doppelgängers.
But first, another pair of duplications!
Right: “Overexpression and biological function of TMEM48 in non-small cell lung carcinoma” (Qiao et al 2016)
A convenient starting point is the curious case of Youbing Xia, who used the unexceptional email account “youbing_x@163.com” as corresponding author of “Analysis on CHENG Dan-an’s exploration of quantification acupuncture and moxibustion stimulation” in 2013. A lengthy hiatus followed, until 2017, when Yongjuan Rui somehow acquired the same email identity and used it (less plausibly) for correspondence regarding the paper “CircRUNX2 through has-miR-203 regulates RUNX2 to prevent osteoporosis”. We note in passing that the loading control from a Figure in that paper has manifested in at least two other locations.
Fig 2D. from CircRUNX2 through has-miR-203 regulates RUNX2 to prevent osteoporosis” (Yin et al 2018)
Fig 9B from “Mir-452-3p: A Potential Tumor Promoter That Targets the CPEB3/EGFR Axis in Human Hepatocellular Carcinoma” (Tang et al 2017).
Then in 2020 the email account was back in the hands of Youbing Xia (or at least a Youbing Xia) for “The Role of Social Support and Environment: The Mediating Effect of College Students’ Psychology and Behavior“. I have so many questions… is Xia aware of the pretender to the name? How do the authors deal with queries intended for the other (such as invitations to respond to a PubPeer thread)? Is this the first step in an elaborate plot to steal Xia’s identity, for nefarious purposes? Is it a social-media counterpart of the Doppelgänger phenomenon that features in literature and popular culture?*
Conceivably Xia felt inadequate in the face of the intricacies of academic publishing, and enlisted a concierge or Dragoman as a proxy for the unfamiliar, foreign-culture manoeuvres of manuscript submission. That hired negotiator opened the email account… then (the speculation continues) the negotiator reused the account for another customer.
The Rise of the Papermills
“Is it possible that through no fault of Zintzaras & Ioannidis, their work was incorporated into a papermill template, accruing hundreds of spurious citations?” – Smut Clyde
This speculation is innocuous and mundane, lacking the intimations of a broader reality that would make it truly satisfying. It does have the advantage, though, of explaining why so many papers by Doppelgänger authors are marked with the stigmata of papermill products, almost as if the hired negotiator also provides papermill services.
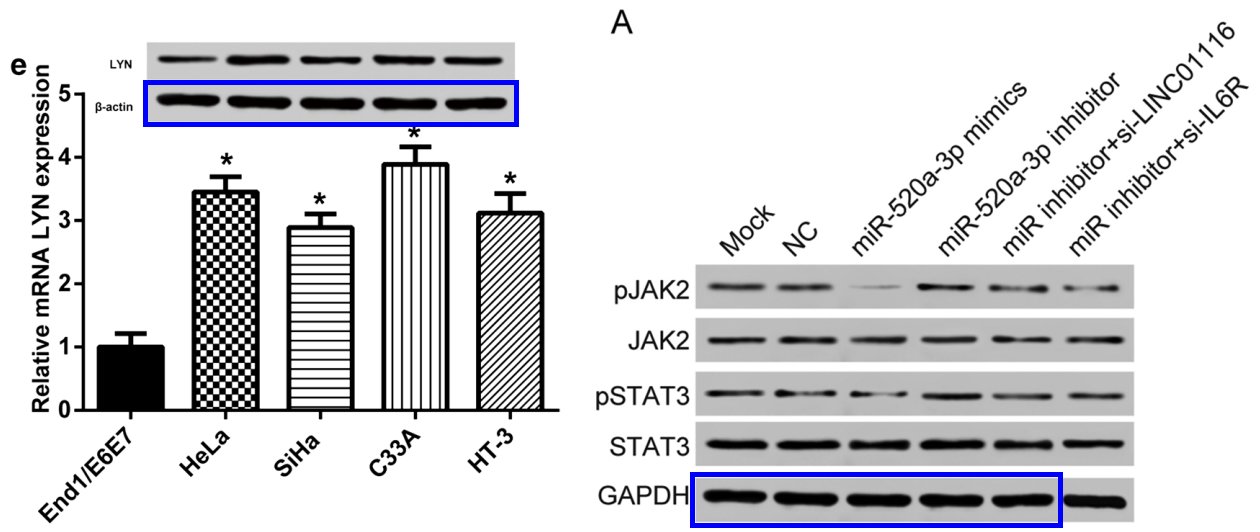
For Xia’s case is not an isolated one. “LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway” (Zhang et al 2018) had two corresponding authors, Lei Ding and Jinlan Jiang. Their e-identities were “doukefengfmmu1@163.com” and “jiangbaoqione@126.com”, where the former had previously been used by Ke-Feng Dou of FMMU, and the latter had been used by Baoqi Jiang. More to come!
[right] Fig 9A from “LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway” (Zhang et al 2018).
The Doppelgänger-adjacent phenomenon of Bilocation is also relevant here. Bilocation – being seen in multiple locations at the same time – was well-regarded in a religious context: classified as miraculous and an excuse for beatification at least. Sadly, it is not such a good thing in Western Blots and flow-cytometry plots, where the general guideline or the aspirational guide is “No more than one paper per Figure”. This may seem unfair but I didn’t make the rules of hagiology.
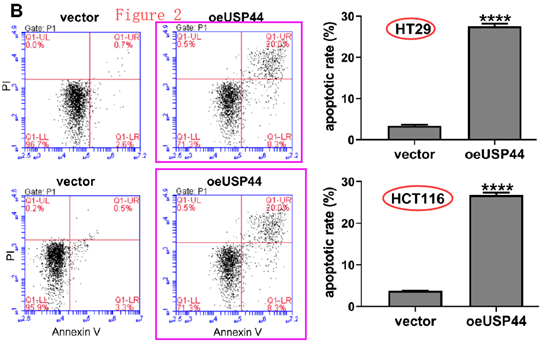
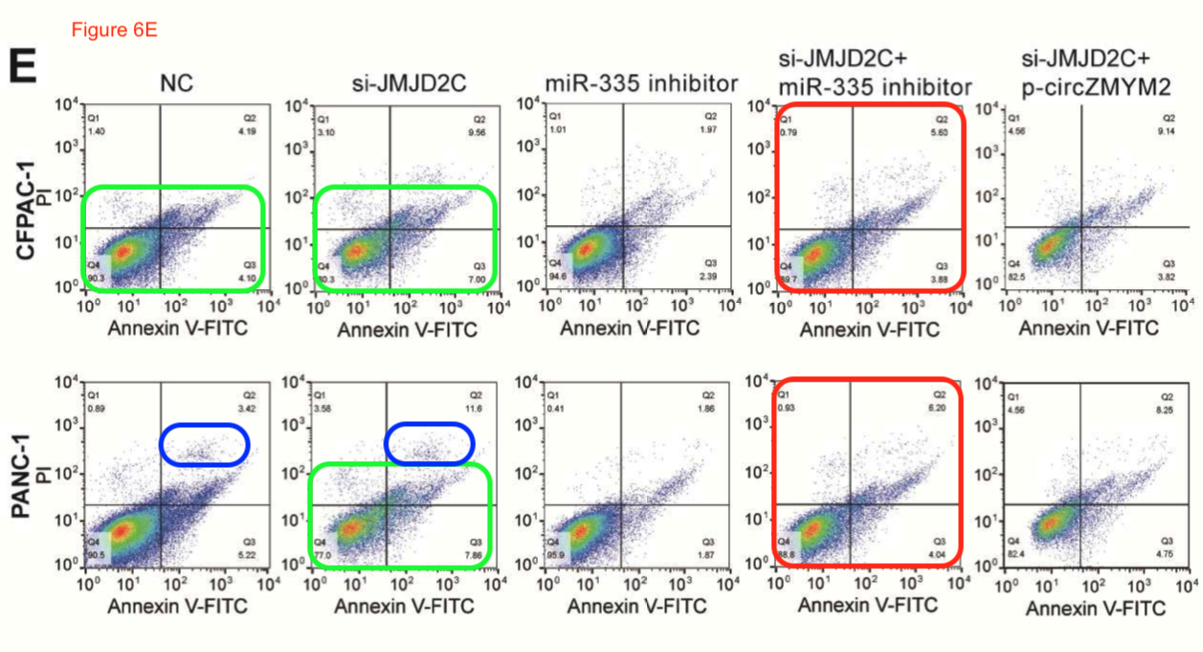
We have already seen the bilocating though unbeatified mouse tumour donors. Here are some bilocating apoptosis-assay scatterplots.
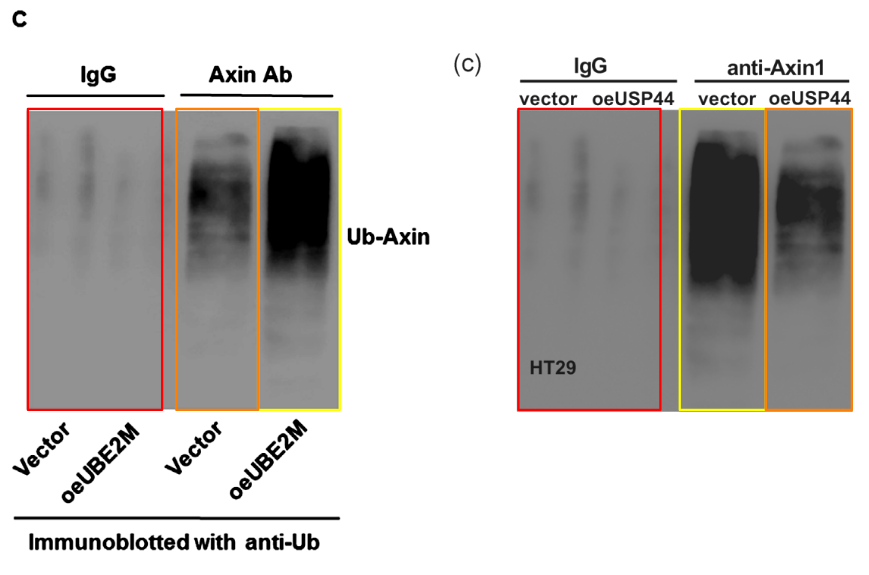
Other figures in that paper also reward further inquiry.
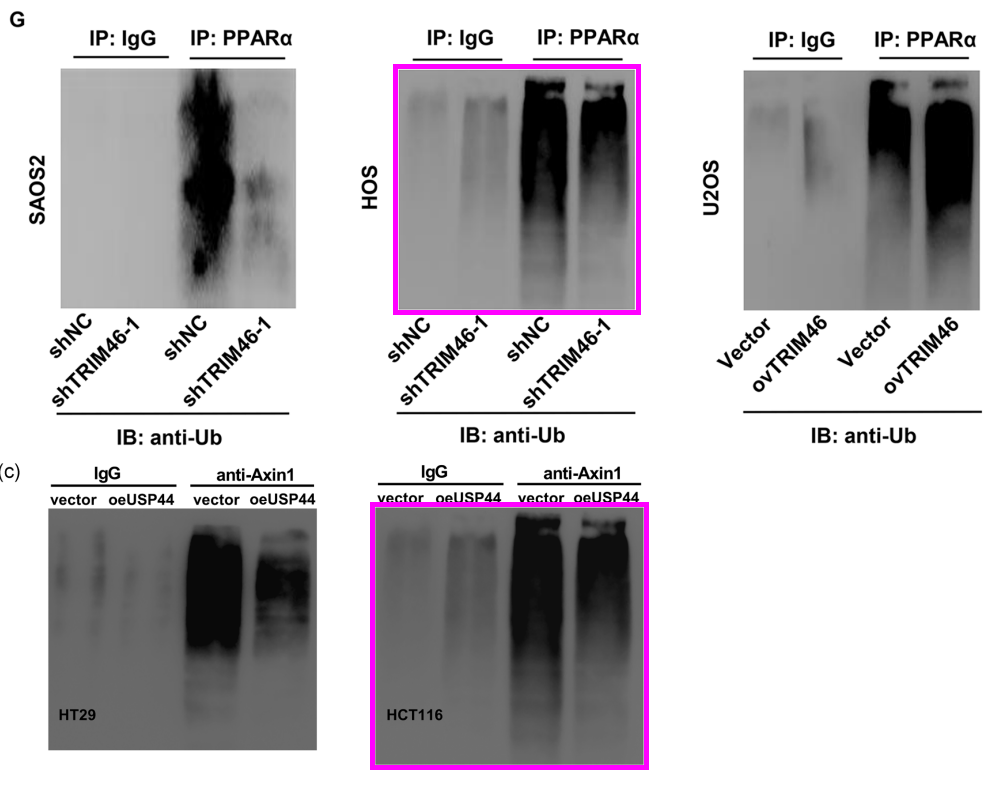
[below] Fig 6c.from Huang et al (2020).
[right] Fig 4C from Huang et al (2020) – HT29 panel.
In fact the multiple manifestations of western blots show up all through the oeuvre I’m describing here. Yes, there is a spreadsheet, up to 220+ papers so far! The earliest entries were a series of 2015 papers. The most recent is “Deleted in liver cancer 1 suppresses the growth of prostate cancer cells through inhibiting Rho-associated protein kinase pathway” (Gong et al 2023), so the suppliers are still active. Names like TRIM29, TRIM65, USP44, FAM46C, FAM84B, ARHGAP10, ARHGAP30 turn up a lot in the titles… I do not know if these are families of cell-signal proteins, or just vanity number-plates. And I must credit ImageTwin for doing much of the work for filling this table.

Fig 3 from “6-Gingerol Attenuates Ischemia-Reperfusion-Induced Cell Apoptosis in Human AC16 Cardiomyocytes through HMGB2-JNK1/2-NF-κB Pathway” (Zhang et al 2019).
Fig 4d from “TRIM27 promotes the development of esophagus cancer via regulating PTEN/AKT signaling pathway” (Ma et al 2018).
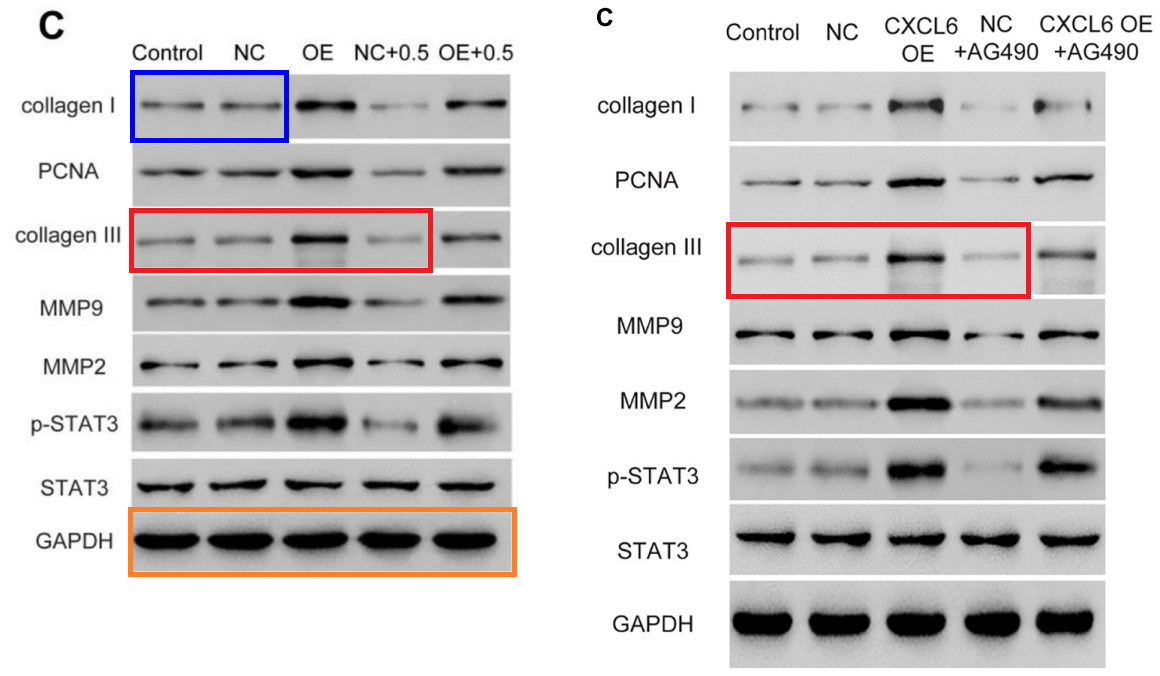
According to Hong Zhang – corresponding author of “ErHuang Formula Improves Renal Fibrosis in Diabetic Nephropathy Rats by Inhibiting CXCL6/JAK/STAT3 Signaling Pathway” (Shen 2020) – the problem sprang from time-share labs:
“The experiments involved in this work were completed from 2014 to 2016. I noticed that at that time I shared the same experimental lab with the authors of “TRIM65 is a potential oncogenic protein via ERK1/2 on Jurkat and Raji cells: a therapeutic target in human lymphoma malignancies” (Wang et al.). We believe some mistakes happened at that time. My paper was published in 2020, there is a long-time gap between the experiments and publication, and the first author has graduated. I am sorry for the mistakes and misuse of the picture. I have contacted with the Editorial office and decided to retract the paper to avoid any further misunderstanding. I will update the future progress timely.“
This accounts for the band fragments generously shared with Wang et al (2018). Though Wang et al. are based in PLA150 Hospital in Luoyang, while Hong Zhang is affiliated to an Institute of Interdisciplinary Integrative Medicine Research within Shanghai University of Traditional Chinese Medicine.** It is unclear why either team would commute 1000 km to work in a laboratory with the other.
Fig 4E and Fig 5C from Shen et al (2020).
Hong Zhang was also senior author of Sun et al (2019), so less speculation is required to explain the band manipulation occurring there.
[right] Fig 6C from “CXCL6 Promotes Renal Interstitial Fibrosis in Diabetic Nephropathy by Activating JAK/STAT3 Signaling Pathway” (Sun et al 2019).
Anyway, this retraction happened, of “Silencing of SOX12 by shRNA suppresses migration, invasion and proliferation of breast cancer cells” (Ding et al 2016):
“This Retraction follows an Expression of Concern relating to this article previously published by Portland Press. This article is being retracted from Bioscience Reports at the request of the authors, following receipt of a notification from a reader alerting the Editorial Office to similarities that exist between the Figure 5C Cyclin/BT474 western blot bands from this article and the Figure 5C B-Carwnin/HO-8910 bands from a paper by Cheng et al. 2015 (doi: 10.18632/oncotarget.4541). During our investigation, the GAPDH bands from figure 3D were also highlighted by a reader as being very similar to those found in Figure 1E from Guo et al. 2017 (https://doi.org/10.1038/s41598-017-01832-y) and Figure 6E of Zheng et al. 2020 (https://doi.org/10.1186/s13046-019-1504-5). The authors were unable to provide the requested original data and responded to our queries by repeating the experiment, providing the Editorial Office with the new data. However, the Editorial Board did not feel that an adequate explanation to the duplicated blots was provided and remain concerned about the lack of original data. The authors therefore wish to retract the article, and the Editor-in-Chief and Editorial Board agree with the retraction.”

[right] Fig 2D from “TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells” (Wang et al 2016).
[middle] Fig 6E from “TRIM6 promotes colorectal cancer cells proliferation and response to thiostrepton by TIS21/FoxM1” (Zheng et al 2020);
[right] Fig 3D (Ding et al 2016).
Ding et al are from Shanghai East Hospital, Tongji University School of Medicine. Guo et al hail from Shanghai Key Laboratory of Female Reproductive Endocrine-Related Disease, Fudan University, Shanghai. We already saw Hong Zhang’s affiliation. Perhaps a theme is emerging. Indeed, the spreadsheet lists eight papers from Fudan University, eight from hospitals affiliated to Shanghai University of Traditional Chinese Medicine (including Zhang’s two), and 30 from hospitals attached to Tongji University.
‘TigerBB8‘ informs me that the structure of hospitals and medical universities in Shanghai is far from centralised: several large universities have acquired their own little empires of hospitals. It seems to be the rule that any hospital needs to have a university in charge. As well as Tongji and the University of TCM, Shanghai Jiao Tong University and the Naval Military University (formerly the Second Military Medical University) have staked out their own manors. Tiger advises me that those last two have bad reputations for junk science (Xuetao Cao rose to be Vice-President of The Second Military Medical University before his further promotion to Mandarin of Research Integrity). Here they have contributed 28 and 10 entries to the spreadsheet, respectively.
The 16 secret retractions of Xuetao Cao at JBC
Chairman Cao’s 16 Retractions They Don’t Want You to Know About
For the same of completeness, I mention Shanghai University of Medicine & Health Sciences, which has an affiliated Shanghai Sixth People’s Hospital East, responsible for four entries. And Nanjing Medical University and its Shanghai General Hospital, responsible for two. These numbers might have missed a few papers with complicated multiple authorship.
So Shanghai is disproportionately represented. What are the other risk factors for illustrative bilocation? That is not a sentence that I expected to find myself writing today, but I prepared a checklist anyway. The more of these risks found in a paper, the better the odds that some of its Western Blots will bilocate into another paper in the same oeuvre (though signed by different authors).
- Authors affiliated to a hospital or university in Shanghai.
- GSEA plots (Gene Set Enrichment Analysis).

- Flow-cytometry apoptosis assays with a family resemblance, like an inverted delta or a mathematician’s Nabla symbol (what the protagonist of Giles Goat-Boy called an escutcheon), often top-heavy and sometimes lop-sided.
- Flow cytometry cell-cycle assays where the fitted contributions from the G0/G2, S and G2/M phases are coded in tasteful pastel shades (in contrast to the usual colour scheme of red humps and a cross-hatched mesa).
- Purple-stained Transwell invasion and migration assays are common, though not specific, as purple staining is standard in this line of research. Adhesion assays are more distinctive.

Fig 5B from “TMEM140 is associated with the prognosis of glioma by promoting cell viability and invasion” (Li et al 2015).
Fig 5 from “Effect of CCL2 siRNA on the cell migration and adhesion in human glioma cell lines U251 and U373” (Lu et al 2017).
My working hypothesis is that somewhere in Shanghai, a private laboratory offers services in biomedical experiments and bioinformational analyses. Its services are popular among medical clinicians who need a foreign-language journal publication on their CVs as a meaningless formality before they can receive promotion, akin to renting an academic gown for a graduation ceremony.
The hypothesis can become a little more specific. Many of the early papers in the corpus credit JRDUN Biotechnology for supplying RNA sequences for transfection, and reagents like RIPA. It seems odd to single out a supplier of such commodity chemicals as Giemsa stain and phosphate-buffered saline solution, but whatever.

JRDUN Biotech even features in Acknowledgements, for “bioinformatics analysis”. What are the odds that they supplied their customers with experiment results as well? A pity about the recycled nature of those results. Sadly, when you outsource the research, you forfeit the opportunity for oversight and quality control, and the material you receive could just be collages, already sold repeatedly to other customers.
I hasten to add that these JRDUN papers, with their GSEA plots and Nablas and pastel-palette cell-cycle histograms, only account for a minority of the entries in the spreadsheet. Other features bilocate across papers: scatterplots modelled after the Nike Swoosh logo. Tumour collections that resemble sprouting chick-peas. Colony plots, stricken with acne. It seems likely that JRDUN Biotech is only one contributor of material within a larger network of outsourced experiment specialists. But it is really a warp and weft of shared Western Blottery that ties most entries into this oeuvre.
[right] Fig 7A from “MircoRNA-1275 promotes proliferation, invasion and migration of glioma cells via SERPINE1” (Wu et al 2018) [retracted].
Figs 2C, 5C from “MiR-130a-3p inhibits the viability, proliferation, invasion, and cell cycle, and promotes apoptosis of nasopharyngeal carcinoma cells by suppressing BACH2 expression” (Chen et al 2017).
Fig 3B from “Up-regulation of miR-10b-3p promotes the progression of hepatocellular carcinoma cells via targeting CMTM5” (Guan et al 2018).
All this charitably assumes that the nominal authors have outsourced only the experimental work that they were not equipped or qualified to perform in person. It might also be that some partner in the network wrote the manuscripts as well.
As is the custom, most of the entries in the spreadsheet are one-time authors (that is to say, clinicians) but a few academics are mixed in as repeat customers. Professor Zhongping Cheng of Tongji University School of Medicine has been to the well ten times… or at least his minions bought first-authorship slots on ten production-line compositions, while the boss graciously accepted Corresponding Author status. An eleventh paper is only suspected of papermill provenance.
Another repeat customer is Dongliang Yan, at Shanghai Sixth People’s Hospital East. Only three papers from this source, though!

To my untutored eye these are convincing high-quality papers when seen in isolation, with little in-your-face internal evidence of falsification. The western blots and flow cytometry results are from actual electrophoresis gels or cell-survival experiments (setting aside the problem that those experiments involved different cell-lines and conditions and protein markers from how they are labelled). So for once I am not accusing the journal editors and peer-reviewers of incompetence and dereliction, nor calling for the literature reviewers who cite these fictions to be cashiered in the scholastic equivalent of a court-martial. That is another sentence I was not expecting to write today.
Alternative Doppelgänger-themed title:
The Magus Zoroaster, my dead child,
Met his own image walking in the garden
As promised, some more email Doppelgängers and their associated WBs:
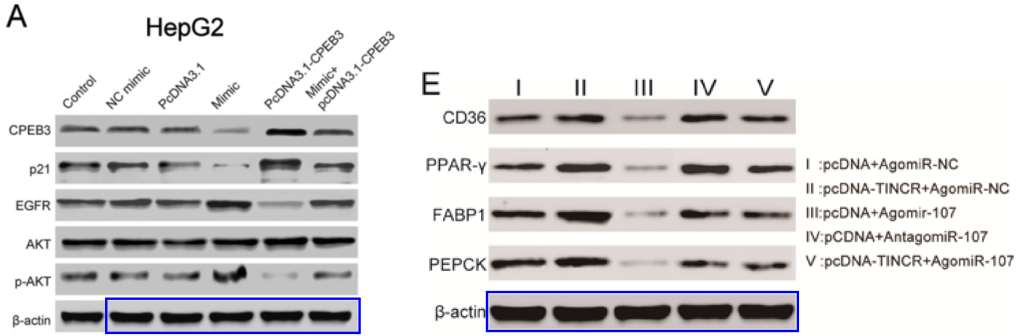
[right] Fig 6E of “LncRNA TINCR/microRNA-107/CD36 regulates cell proliferation and apoptosis in colorectal cancer via PPAR signaling pathway based on bioinformatics analysis” (Zhang et al 2019),
The corresponding author of Zhang et al (2019) was Lianfeng Zhang, who for some reason chose “ssmu_pan@163.com” as an email identity. It was previously used in 2008 by Yi-Qing Pan, from Shanghai Jiao Tong University School of Medicine. For Tang et al (2017), the email identity used by corresponding author Wei Meng was “xiao_yue51833@126.com”. This appears to be passed around a lot, and was previously used in 2008 by a Shi-Yue Su.

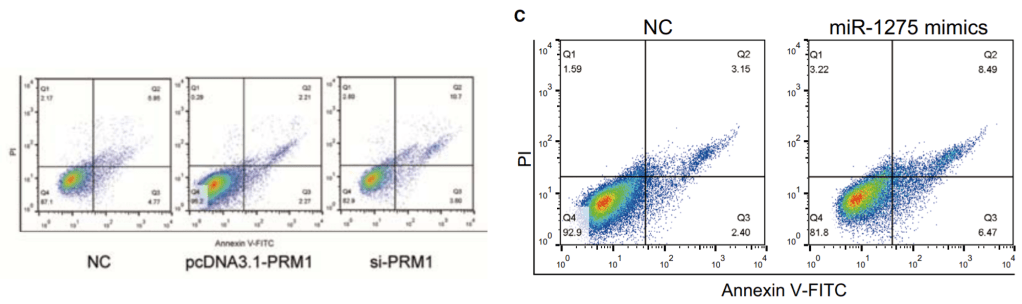
[right] Fig 1D from “Impact of protamine I on colon cancer proliferation, invasion, migration, diagnosis and prognosis” (Chen et al 2018).
The corresponding author of Chen et al (2018) was Ye Feng, using the email address “xue_zhanggang@126.com”.

[right] Fig 2A from “TMEM49-related apoptosis and metastasis in ovarian cancer and regulated cell death” (Zheng et al 2016).

[right] Fig 2A from “Knockdown of EHF inhibited the proliferation, invasion and tumorigenesis of ovarian cancer cells” (Cheng et al 2016).
[right] Fig 7B from “MircoRNA‐1275 promotes proliferation, invasion and migration of glioma cells via SERPINE1” (Wu et al 2018b) [retracted].
Here the corresponding author of Wu et al (2018a) was Jinchun Xing, who chose the email identity “xibuliadi@163.com” for correspondence. That account was previously associated with Xiao-ying Wu who used it with a 2003 manuscript before it fell into abeyance and desuetude (which might be the same thing).
“We are extremely guilty and distressed”
“Now no-one wants ForBetterScience to become an all-Papermill channel. And we cannot really expect to shame or inspire scriveners in the academic-ghostwriter industry to seek out more constructive applications for their talents, so the point of exposing them is not immediately obvious.” -Smut Clyde
First author of Wu et al (2018b) was one Dong-Mei Wu of Jiangsu Normal University: #17 on the Retraction Watch leaderboard with 35 retracted papers (many of them from purchased from the ‘Contractor papermill’), and recipient of repeated castigation from the National Natural Science Foundation of China. Within the present oeuvre, also author of “Long noncoding RNA nuclear enriched abundant transcript 1 impacts cell proliferation, invasion, and migration of glioma through regulating miR-139-5p/ CDK6” (Wu et al 2019).
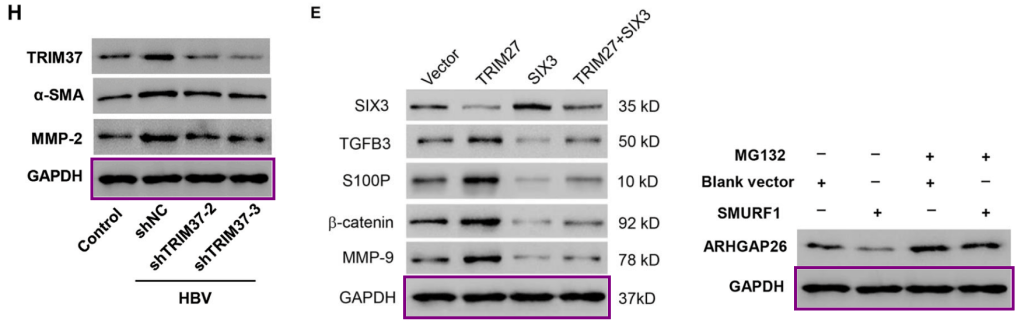
[middle] Fig 4E from “TRIM27 acts as an oncogene and regulates cell proliferation and metastasis in non-small cell lung cancer through SIX3-β-catenin signaling” (Liu et al 2020).
[right] Fig 6D from “SMURF1-mediated ubiquitination of ARHGAP26 promotes ovarian cancer cell invasion and migration” (Chen et al 2019).
[right] Fig 2B from “Inhibition of Proliferation, Migration, and Invasion by Knockdown of Pyruvate Kinase-M2 (PKM2) in Ovarian Cancer SKOV3 and OVCAR3 Cells” (Miao et al 2016).
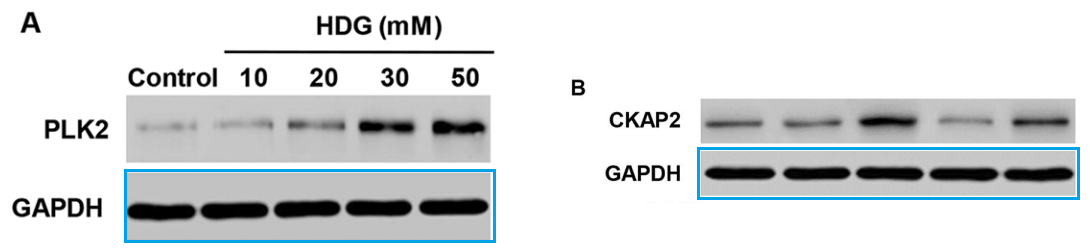
Now a seven-fold multiplication! Beat that, St. Padre Pio!
[right] Fig 6B from “Involvement of FAK-ERK2 signaling pathway in CKAP2-induced proliferation and motility in cervical carcinoma cell lines” (Guo et al 2016).
[right] Fig 8c from “Advanced glycation end products induce the apoptosis of and inflammation in mouse podocytes through CXCL9-mediated JAK2/STAT3 pathway activation” (Yu et al 2017).
Finally, some bonus “Swoosh” scatter-plots.

Two corresponding authors here, Jixue Zhao and Yaozhong Zhang, who chose the email identities “czw_ahmu@126.com” and “czyansd@126.com”. These were previously and more plausibly used by Zhi-Wu Chen in 2007 and C-Z Yan in 2010.
And since I’ve already mentioned Chen et al (2018) and Wu et al (2018b), here are their Figs 3D and 4C.
* Not to forget a fine essay by MacDonald Critchley about heautoscopy and Doppelgänger encounters as a sign of neurological malfunction.
** Also the alma mater of Junyi Shen.

Donate to Smut Clyde!
If you liked Smut Clyde’s work, you can leave here a small tip of 10 NZD (USD 7). Or several of small tips, just increase the amount as you like (2x=NZD 20; 5x=NZD 50). Your donation will go straight to Smut Clyde’s beer fund.
NZ$10.00











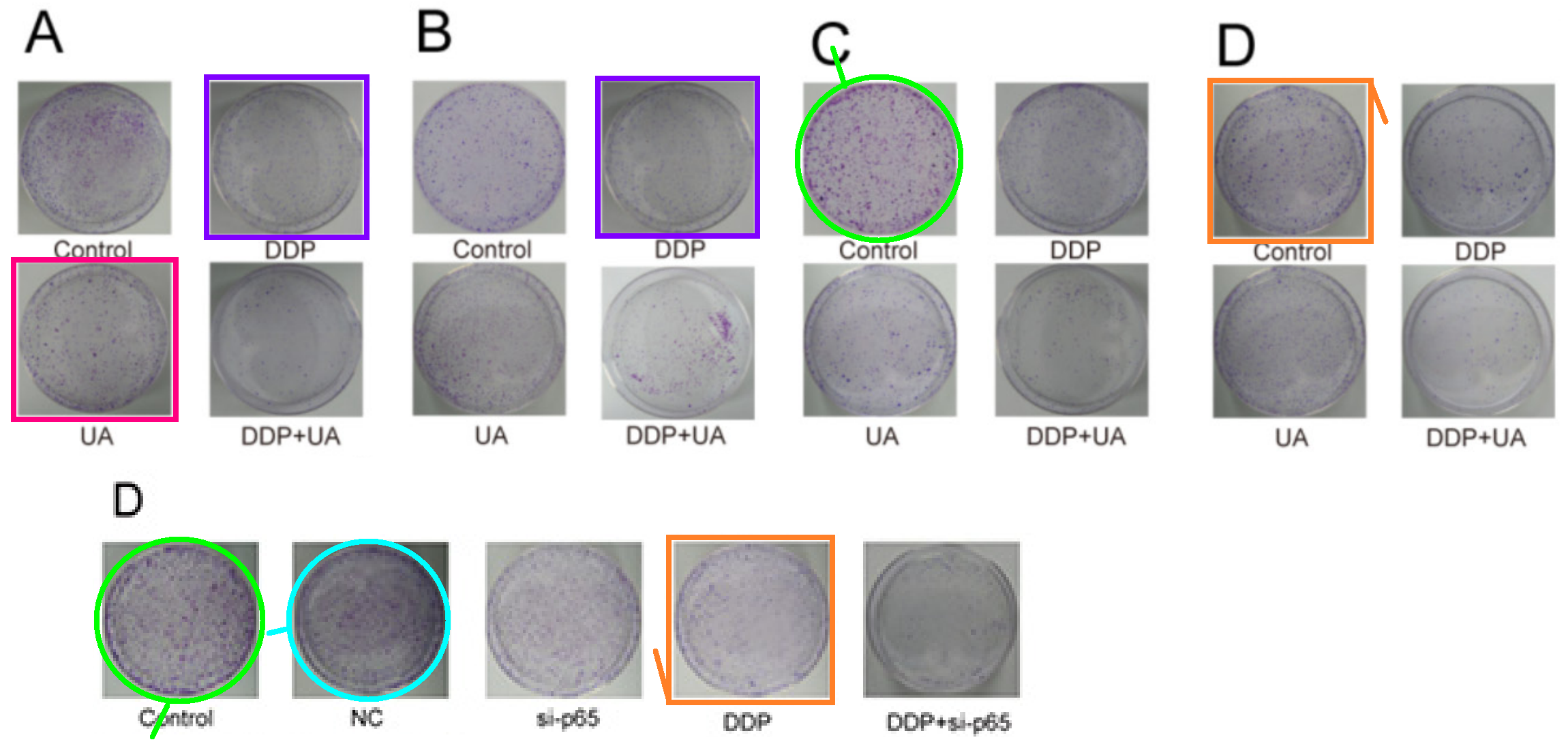
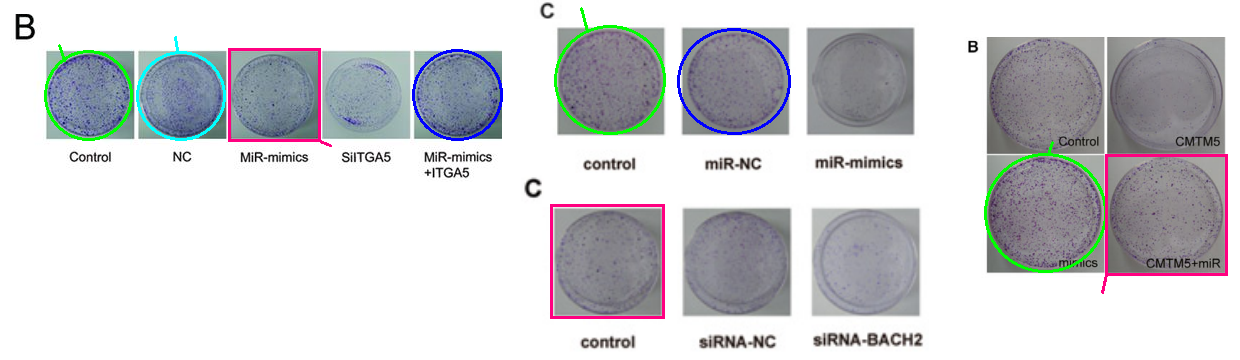
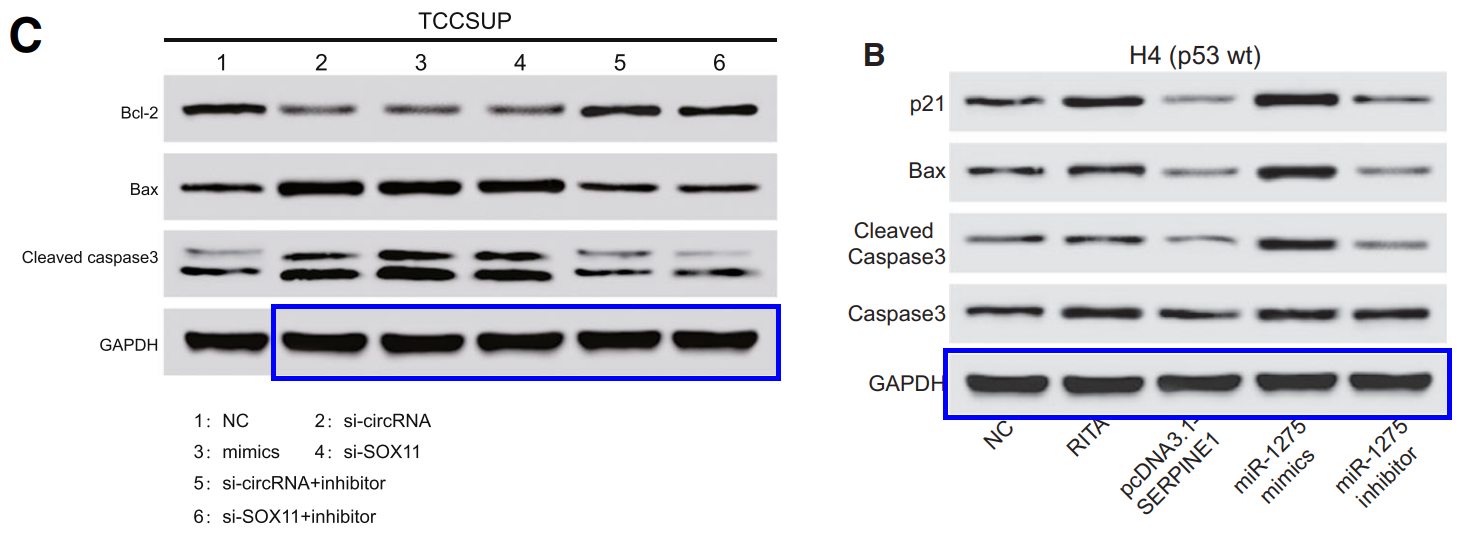

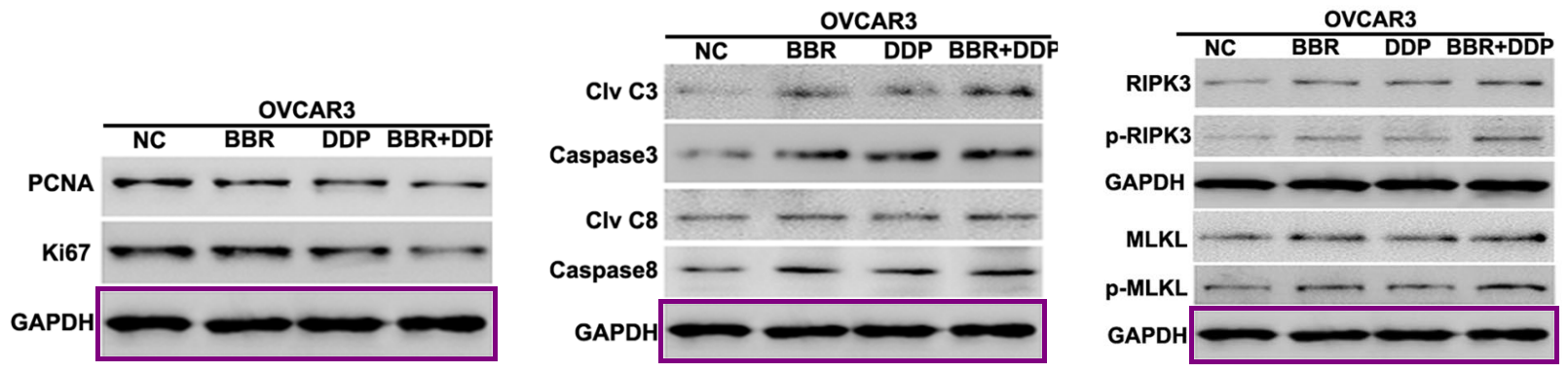



The discovery of papermills deserves a Nobel Prize. The process has been a monumental piece of work for the benefit of science and humanity.
LikeLiked by 1 person
Splendid work. The presence of just one papermilled entry from 2022 and one from 2023 could suggest this papermill has run out. If so, I fear a new one will soon be established.
LikeLike
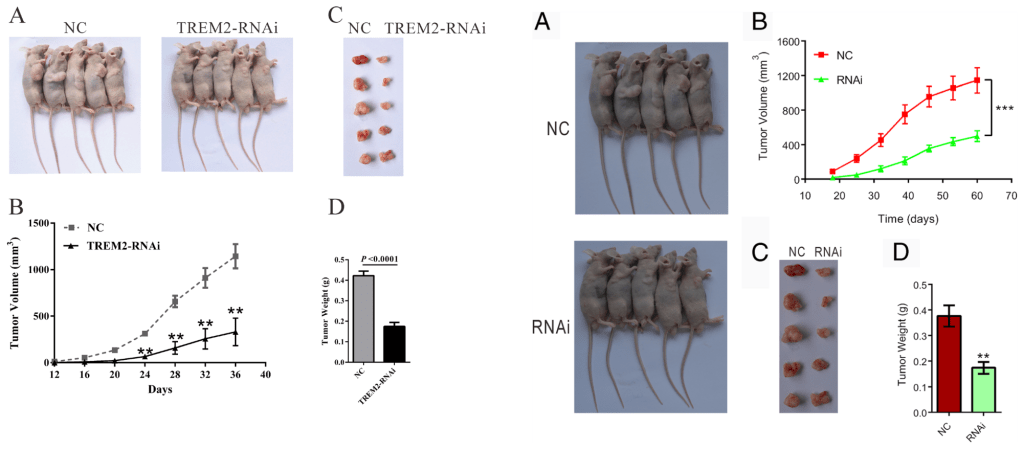
A blog I hadn’t previously seen. The tumors I do recognise though…
LikeLike