This is a call to action, or rather to inaction, to protest against the pervasiveness and acceptance of research misconduct.
I invite you to not do certain things:
- Do not cite papers by dishonest scientists
- Do not go to their seminars on your campus
- Do not go to any conferences where they feature as speakers
It is act act of passive resistance and will bear absolutely no consequences upon your academic career. It will only be noticed when suddenly seminar audiences are largely empty, or when a conference cannot take place because you and others refused to pay huge fees to watch an invited fraudster on stage.

Conference podium presence is what distinguishes science stars from science precariat, an invitation to speak at a conference is a sign of functional networks, peer acceptance and admiration for your scientific achievements. This is why some academics resort to predatory conferences, to fake such recognition on their CV, where one is lacking. Others are stars, much sought after, up until their trip over their own dishonesty, and then the invitations all but stop happening. First thing every scientist tainted by misconduct findings does, is to try to get invited again to meetings and workshops, like the plant scientist Olivier Voinnet and the diabetes researcher Katrin Maedler tried, but so far failed. Some are more successful, like the cancer and ageing researcher Karl Lenhard Rudolph, whose example will be discussed below.

I suggest you don’t go, neither as audience nor as speaker. If you like, you can send an (anonymous or signed) email to the event organisers, explaining why you are not coming. You might think it is unfair towards other invited speakers, but it is not. They are not supposed to be there either, at least if they care about research integrity. Hence: withdraw your participation if you are also an invited speaker, do not help a cheater by association with you on the stage or at a lavish social event. Maybe the conference organisers will even un-invite the cheater, and quietly replace their name on the programme with someone more deserving – such things did happen actually.
How to spot a Zombie Scientist
Official misconduct findings should be the final sign for you to know whcih cheaters to avoid. These zombie scientists have no place in science, there is no way to reintegrate them, they cannot be trusted with research ever again. They will never become honest, but they will design new tricks to avoid getting caught again, while still producing unreliable shady data. They will continue to spawn next generations of cheaters in their labs, while punishing those “trainees” who want to do good science. Once these fallen giants get their power back, they will take revenge on every whistleblower and critic in their academic community.
If someone officially guilty of fraud speaks at a conference, it’s because the organisers wish to help that fallen cheater rise again, and you will play a key part in that scheme. Resist passively, all you have to do is not to go. Saves money even, and is good for the environment and climate.
What other signs whom to avoid? Retractions can be a sign, but only in context. There are scientists who retract their own papers after having found out that a lab member falsified the data. The former should be commended and supported, while the latter should be rightly avoided as science fraudsters, even though their both names are on the same retraction.
Then again, retractions, or the absence thereof, are often merely a sign for two things:
- How many friends a fraudster has lost: the more retractions, the fewer friends remain
- Specific journal ethics: fraudulent papers in Journal of Biological Chemistry (JBC) get retracted en masse, while Cell Press cannot care less. Also, some academic chief editors don’t care about research misconduct at all, some actively engage in it themselves.
A clever cheater simply avoids certain journals. This is exactly why K Lenhard Rudolph never retracted a paper, he even managed to evade some corrections. Absence of retractions certainly does not imply honesty.
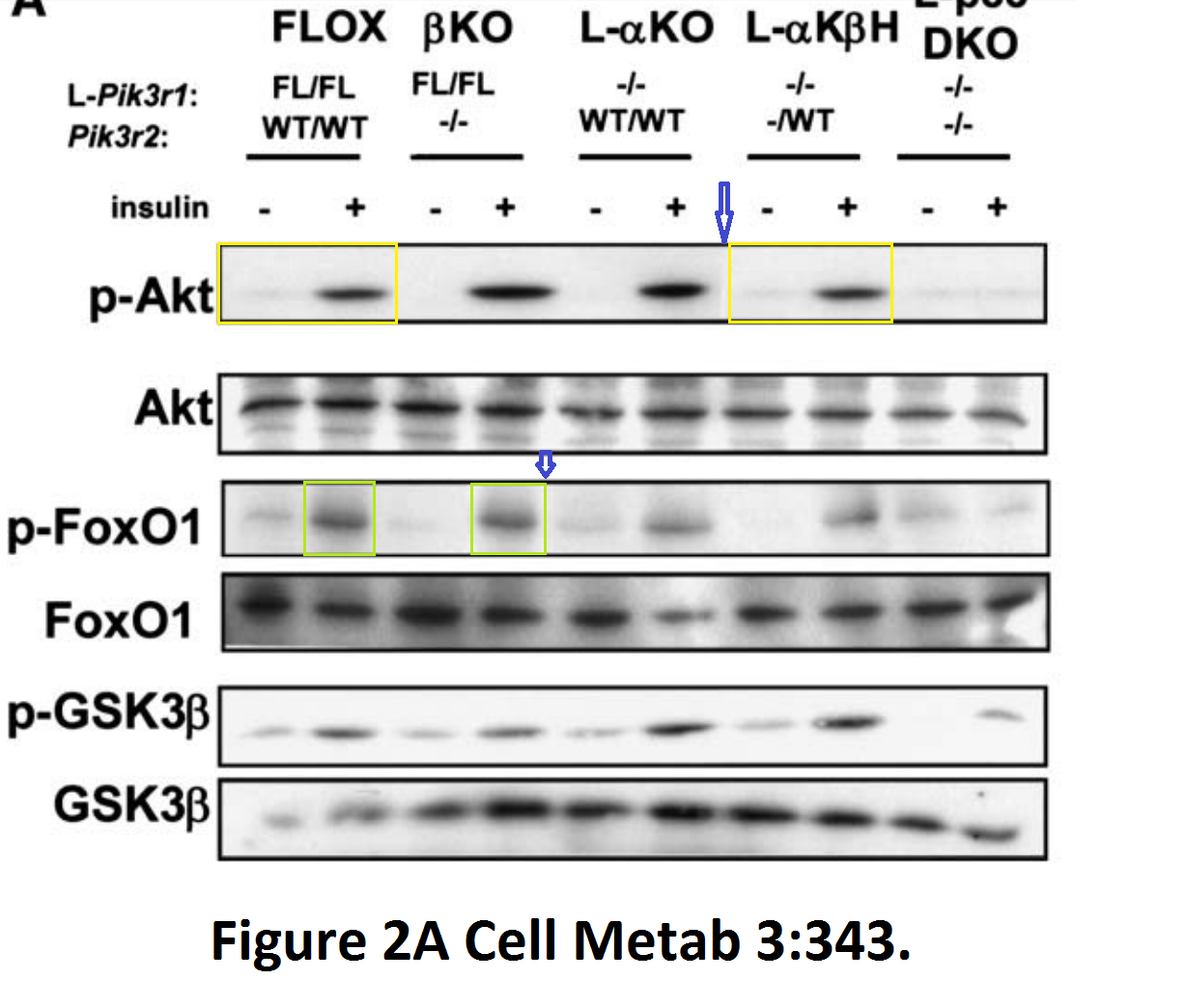
PubPeer.com is a key source of reference, but of course you should look critically at the available evidence and the recurrence of certain author names, and then make an informed decision. It should not matter if that scientist is an absolute star, adored by everyone like Carl Ronald Kahn is. This diabetes researcher from Harvard has 40 papers on PubPeer, many with very serious issues. And yet Kahn decided to ignore all evidence and not to do anything, at least unless the journals make him. Such refusal to act on available evidence of manipulated data was defined elsewhere as research misconduct in itself.

Kahn has 3 retractions, two in JBC, and one in Journal of Clinical Investigation. None of that damaged his reputation in any way, Kahn is a celebrated awardee, and a welcome seminar guest and keynote speaker at many conferences. Are you planning to come?
We stand with Rudolph!
Now I would like to present you an bizarre “We Stand with Rudolph” conference at his Fritz-Lippmann-Institute (FLI) in Jena, the second instalment of a travesty event which you definitely should avoid. Not just that, avoid all events these organisers and keynote speakers appear in, and be careful with other participants as well. They made their stand of research integrity, not in a good way, now do you want to be their admiring audience?

Rudolph, then FLI scientific director, was found guilty of research misconduct twice, first by his own Leibniz Society (which operates the FLI in Jena) and by the Germany Research Council, the DFG, which barred the telomere researcher from funding for 2 years (read here and here). Prior to those misconduct findings, Rudolph’s activities caused a police raid on FLI because of animal abuse – mice were experimented upon without ethical approval. FLI was then banned from performing any animal experiments whatsoever.
Getting into media with research animal abuse and even causing a police raid is the worst thing a German scientist can do. Maybe this is why Rudolph was so thoroughly investigated, on whistleblower evidence which is even now not available on PubPeer or anywhere else. Prior to that, Rudolph was the absolute star of German and international cancer and ageing research, recipient of €2.5mn heavy ERC and Leibniz awards, among other things.
After the misconduct findings, the condition for FLI to receive any future funding from the Leibniz Society was that Rudolph steps down as the institute’s scientific director, which he promptly did. He also resigned as that year’s president of the German Stem Cell Network.
But Rudolph remained professor and senior group leader at FLI, apparently the institute and the affiliated University of Jena were loath to lose Rudolph’s creative approach to impactful science. As I learned, the university authorities even failed to offer his PhD students an opportunity to change mentor, so that these had to continue their PhD training supervised by someone found guilty of research misconduct.
Despite his catastrophic fall and public humiliation, Rudolph certainly did not want to lose connection to his peer community. He almost spoke as invited speaker at a Keystone symposium and the ISSCR annual meeting in 2018, but somehow, without any explanation or announcement, someone unrelated was given his slot (more here). In December 2018, the German Society for Ageing Research (Rudolph used to be its president, 2008-2014) had its annual conference at FLI in Jena, for some reason the programme for exactly that event is secret, while the society’s website pretends it never happened.
In March 2019 however, Rudolph was chairing a session at Gordon Research Conference in California, organised by Amy Wagers and Cedric Blanpain. Now, Wagers is a Harvard professor, known for her “young blood rejuvenation” parabiosis research, where she had to retract two papers, one in Nature, one in Blood, while her postdoc took the full blame. Even if you ignore the data manipulation: Wager’s rejuvenation discoveries were soon debunked by other scientists and even by the FDA, but it did not hurt her career in the long run, being a protegee of the mighty Irving Weissman. You now probably see why Wagers saw it as her duty to support Rudolph in difficult times.
If his peers don’t invite Rudolph often enough, he invites them. Even if the Leibniz Society and the university refuse funding, his own FLI is more than happy to divert research funds for this occasion, as I found out before. And the friends sure come, last year’s “We Stand with Rudolph” event was apparently such a success, that Rudolph and his FLI colleague Hellen Morrison (this time with Alessandro Ori) announced another one for 2020 (archived version, just in case, here). As keynote speakers are listed Andrew Dillin, Linda Partridge and Mike Stratton, the latter is the Sanger Institute director responsible for the scandal of misuse of African DNA, who also was whitewashed of bullying accusations after whistleblower complaints were dismissed. All these speakers know exactly why they are invited, and this is why they and others will be coming to stand with Rudolph.
And now FLI had another genius idea how to reform Rudolph and other cheaters at the institute. As reported in Nature, Rudolph’s science will be now checked by research integrity professionals:
“The FLI’s head of core facilities, Matthias Görlach, had a less conventional idea. He contacted Enrico Bucci, a molecular biologist who had visited the FLI for some PhD work 18 years earlier, and with whom he’d kept in touch. Bucci was now in the business of checking research papers, Görlach knew; in 2016, he’d founded a science-integrity firm called Resis, based in Samone, Italy. Could the company perhaps help the institute to avoid errors in future?
So began a remarkable system of outside vetting, in which researchers at the FLI must send every paper and master’s thesis across to Resis for screening before they submit them for publication.”
Maybe some particularly dim or lazy master students will get caught on plagiarism now. But there is no reason to assume the dishonest FLI scientists intend to get caught again on same tricks, so it is not clear what the new scrutiny is supposed to achieve here. There are so many ways to rig data which leave no traces whatsoever in the final figures: re-labelled gels, loading control libraries, brightness-adjusted images, removed “outlier” data points, discarded mice and patient samples, and never mind all the digital outputs like quantitative real-time PCR, microarrays or mass-spectrometry where the data is just an Excel sheet beckoning to be toyed with.
What the new FLI policy will achieve though, is a stamp of approval for Rudolph’s new science, shiny, spotless and sparkling clean. No manipulations there whatsoever. The elite journals will be queuing again to publish Rudolph’s research, they actually already do, see for example Chen et al JEM 2019. And you will be in the conference audience, listening to Rudolph’s keynote address.
Unless you decide not to go.

Not a single retraction!
Finally, the list of Rudolph papers which the Leibniz Society investigated, updated with the current status. The society’s PR person refused to give me that list, but they gave it to Retraction Watch, where it was published. The Leibniz investigation found research misconduct in 8 of these 11 papers, but the list did not specify which ones. At least it indicated for all eleven papers which figures exactly were flagged. A follow-up investigation by the DFG found misconduct in 3 papers (numbers 4, 6 and 8), incidentally 8+3=11. One of these three artworks, Missios et al Nature Comm 2014, was never corrected, even though DFG declared in December 2017: “Mr Rudolph offered to journal an Erratum.” Seems the journal politely declined!
Elsewhere however, Rudolph had to issue a mega-correction to a 2016 Nature paper, and that one was officially never investigated by the DFG or Leibniz Society! It was much celebrated by FLI though.
- S Schwörer, F Becker, C Feller, AH Baig, U Köber, H Henze, JM Kraus, B Xin, A Lechel, DB Lipka, CS Varghese, M Schmidt, R Rohs, R Aebersold, KL Medina, HA Kestler, F Neri, J von Maltzahn, S Tümpel & KL Rudolph Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals Nature volume 572, pagesE11–E15(2019) Correction 30.07.2019:
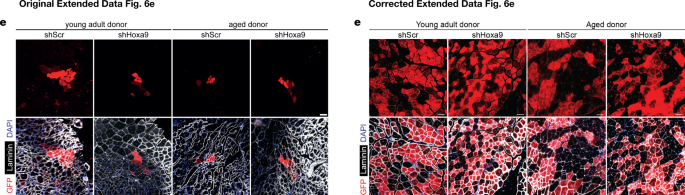
“In this Letter, errors occurred in the following figures. In Extended Data Fig. 6e, the ‘shScr, Aged donor’ image is a duplicate of the ‘Vehicle, Aged donor’ image in Fig. 3f. The images in Extended Data Fig. 6e represent differences in engraftment levels under four experimental conditions; however, these reflect the lower end of the observed overall engraftment rate in the experiment. Figure 1 of this Amendment shows the corrected panels for Extended Data Fig. 6e, with images from the original experiment that best reflect the differences in, and the overall level of, the engraftment rates between the conditions under study (the original images from Extended Data Fig. 6e are shown for comparison).
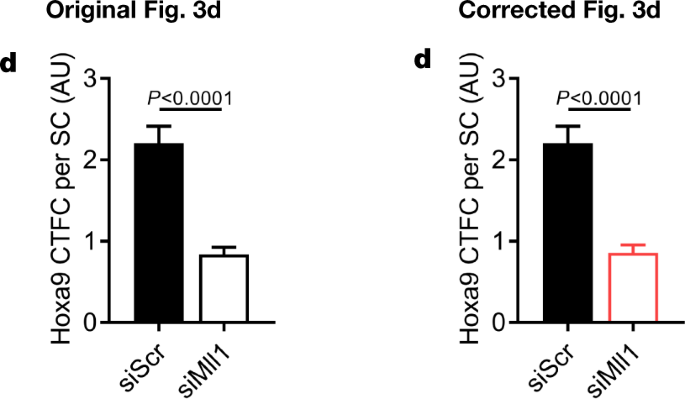
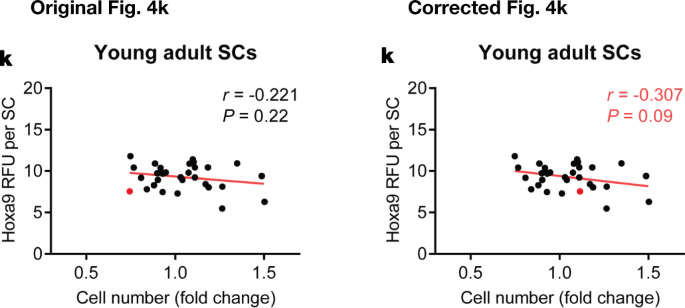
In addition, there are errors in the Source Data for Figs. 3d, 4k, Extended Data Figs. 4f–h, 7f, s, t and 9m–o, q–s due to copy-and-paste errors or due to the presentation of controls that were used for the calculation of P values or error bars shown in the figures. One value that was identified as an outlier in Extended Data Fig. 10g was not labelled as such in the original Source Data and was erronously included for graphical depiction. See Supplementary Information to this Amendment for the corrected Source Data files, and Figs. 2, 3 and 4 of this Amendment for the corrected and original panels for Figs. 3d, 4k and Extended Data Fig. 4f, g, respectively. For the calculation of the P value in Extended Data Fig. 6b, we applied a one-sided paired ratio Student’s t-test (not, as stated, a two-sided Student’s t-test).
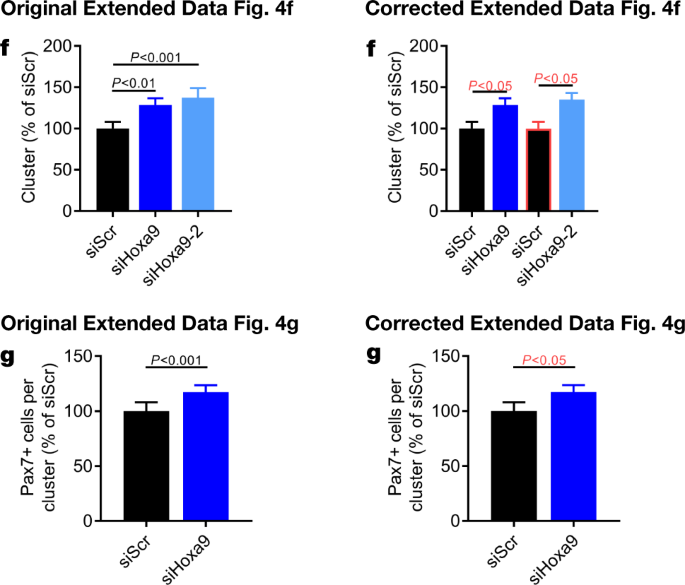
In addition, in Extended Data Fig. 7m, n, p, r, 9i, q–s and 10d, e of the original Letter, errors occurred in data scaling that affect the calculation of P values and the graphical presentation of the data. See Figs. 5, 6 and 7 of this Amendment for the corrected and incorrect panels for Extended Data Fig. 7f, m, n, p–t, 9i, m, q–s and 10d, e, g, respectively, and Supplementary Information to this Amendment for the corrected Source Data. The errors in data scaling occurred because two methods of data scaling were used throughout the study. In some experiments, data of the experimental groups were scaled to the average of the control values; in other experiments, data of the experimental groups were scaled to each of the corresponding controls of a biological repeat, set to 1 or 100. Although both methods of scaling are valid, they should not be combined within one experiment, which happened in the aforementioned figures. This has now been corrected and we include a detailed description of our scaling approach in the Supplementary Information to this Amendment.
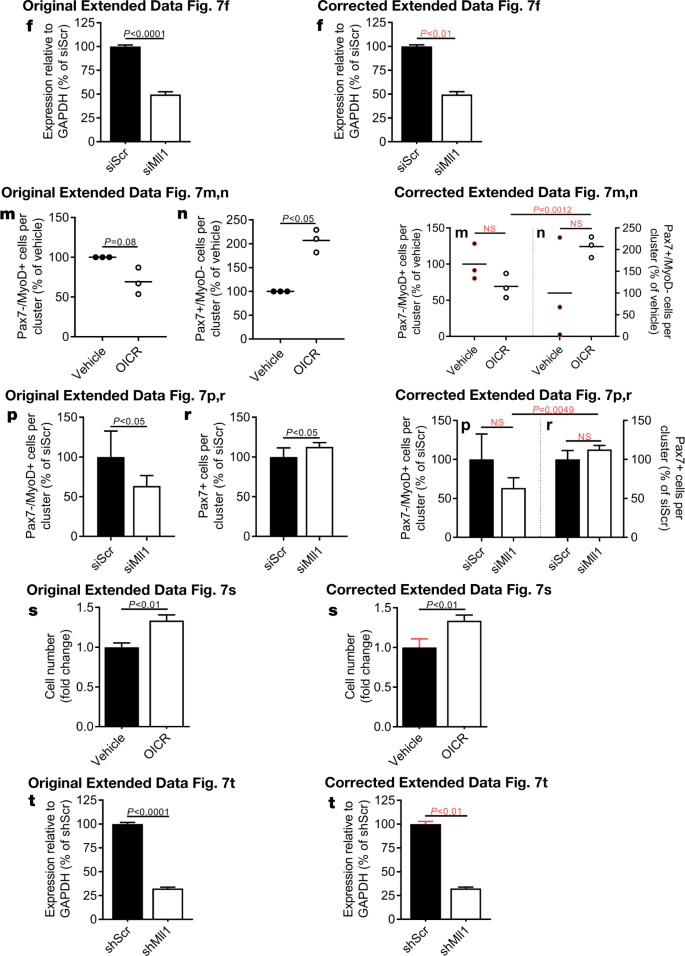
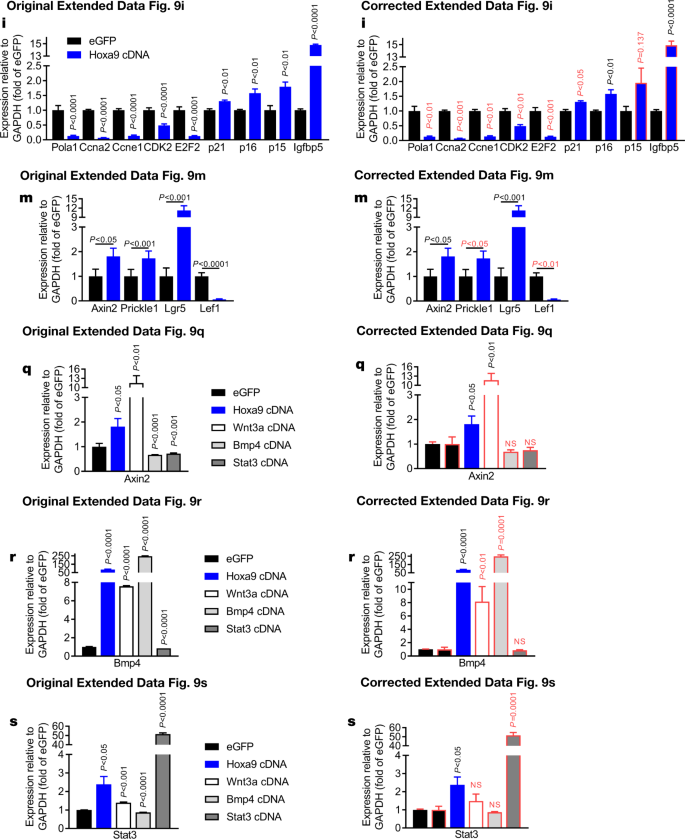
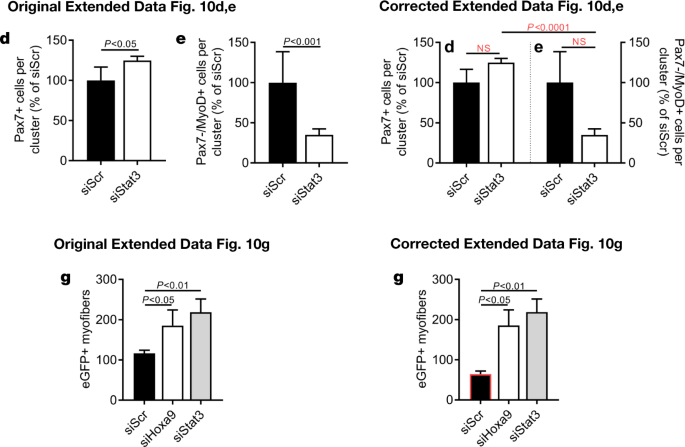
The outlined corrections do not change the conclusions of the original Letter, and we apologize for any confusion that these errors may have caused. The original Letter has not been corrected.“
But now, the Leibniz list. I also added hyperlinks and corrigenda where these happened. Rudolph never had to retract a paper, and he most likely never will.
- Ranganathan V, Heine WF, Ciccone DN, Rudolph KL, Wu X, Chang S, Hai H, Ahearn IM,Livingston DM, Resnick I, Rosen F, Seemanova E, Jarolim P, DePinho RA, Weaver DT. Rescue of a telomere length defect of Nijmegen breakage syndrome cells requires NBS and telomerase catalytic subunit. Current Biology 2001; 11:962-6, Figure 2B.
- Wirth T, Kühnel F, Fleischmann-Mundt B, Woller N, Djojosubroto M, Rudolph KL, Manns M, Zender L, Kubicka S. Telomerase-dependent virotherapy overcomes resistance of hepatocellular carcinomas against chemotherapy and tumor necrosis factor-related apoptosis-inducing ligand by elimination of Mcl-1. Cancer Research 2005; 65:7393-402, Figure 3B and 3C. Correction 15.10.2018
“In the original version of this article (1), in Fig. 3, the images of the cell lines Hep3B and Huh7 infected with either Ad-wt or hTERT-Ad were used twice (top rows of panels B and C) because only one representative picture was taken of the infected cells. The figure legend has been edited in the latest online PDF version of the article to explain the intentional display of the same images. The authors regret this error.”
- Jiang H, Schiffer E, Song Z, Wang J, Zürbig P, Thedieck K, Moes S, Bantel H, Saal N, Jantos J,Brecht M, Jenö P, Hall MN, Hager K, Manns MP, Hecker H, Ganser A, Döhner K, Bartke A,Meissner C, Mischak H, Ju Z, Rudolph KL. Proteins induced by telomere dysfunction and DNA damage represent biomarkers of human aging and disease. Proc Natl Acad Sci U S A 2008; 105:11299-304, Figure 3G. Erratum 10.04.2018
“The authors wish to note the following: “For Figs. 1B, 3B, and 3G and Figs. S2A and S5, the figure legends should have noted that the expression control of GAPDH was run in parallel on separate gels using the same lysates and loading. We apologize for not making this explicit in the figures.”
- Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A, Hildner K, Guachalla LM, Gompf A,Hartmann D, Schambach A, Wuestefeld T, Dauch D, Schrezenmeier H, Hofmann WK, Nakauchi H, Ju Z, Kestler HA, Zender L, Rudolph KL. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 2012; 148:1001-14; Figure 3C and 4C. Erratum 11.09.2014 (hidden by Cell):
“It has come to our attention that the actin loading control for the BATF western blots depicted in Figures 3C and 4C of the article above are from different exposures of the same western blot. Although appropriate actin controls were performed for the experiments presented in Figures 3C and 4C, because the error occurred at the stage of data collection, we cannot provide an image of the correct control bands for the data presented in 4C. The magnitude and impact of this error do not affect the published conclusions. We apologize for any confusion the error may have caused.”
- Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y, Lechel A, Rudolph KL, Langer R, Slotta-Huspenina J, Bader FG, Prazeres da Costa O, Neurath MF, Meining A,Kirchner T, Greten FR. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 2013;23:93-106, Figures 3C and 4C.
- Missios P, Zhou Y, Guachalla LM, von Figura G, Wegner A, Chakkarappan SR, Binz T, Gompf A, Hartleben G, Burkhalter MD, Wulff V, Günes C, Sattler RW, Song Z, Illig T, Klaus S, Böhm BO, Wenz T, Hiller K, Rudolph KL. Glucose substitution prolongs maintenance of energy homeostasis and lifespan of telomere dysfunctional mice. Nature Communications 2014; 5:4924, Figures 5E, Suppl. 5A, Suppl. 5B and Suppl. 5C.
- Wang J, Lu X, Sakk V, Klein CA, Rudolph KL. Senescence and apoptosis block hematopoietic activation of quiescent hematopoietic stem cells with short telomeres. Blood. 2014 Nov 20;124(22):3237-40. Figures 1C and 2C.
- Meena JK, Cerutti A, Beichler C, Morita Y, Bruhn C, Kumar M, Kraus JM, Speicher MR, WangZQ, Kestler HA, d’Adda di Fagagna F, Günes C, Rudolph KL. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 2015 May 12;34(10):1371-84, Figures 2B und 1D. Correction 2.10.2017:
“The authors state that errors occurred during formatting of Fig 2B and Supplementary Fig 3C during the submission process of the original manuscript. The histogram of Fig 2B was mistakenly duplicated in Supplementary Fig S3C. The corrected histogram of Supplementary Fig S3C is shown below. In addition, Fig 2B contains two labeling errors of P‐values, originating from mistakes in the labeling during the graphical assembly of the figure: The P‐value for OSBPL3‐shRNA vs. scrambled shRNA should read 0.0218 instead of 0.0216, and the P‐value for GJB3‐shRNA vs. scrambled shRNA should read 0.0011 instead of 0.0012. The corrected Fig 2B is shown below. For the experiments shown in Fig 2B and Supplementary Fig S3C, a total number of 74–174 cells was analyzed in 3–14 vision fields per cell line.
Further, γH2AX staining of IMR90 cells mentioned in the results section (page 1373, last sentence “… telomerase‐negative BJ and IMR90 fibroblasts exhibited a strong accumulation of DNA damage (staining positive for both γH2AX and 53BP1) at early passage after shRNA transduction (Fig 2A, C and D, Supplementary Fig S3A and B)…” was not depicted in the paper, and the 53BP1 staining was performed only on the BJ cells. To correct for this mistake, the text and data presentation of the result section on the description of DNA damage foci (page 1373, last sentence, and page 1374, first paragraph) is specified as follows: “The experiments on IMR90 cells were conducted only for γH2AX staining. These results are now provided in Supplementary Fig S8”.
The description and presentation of Western blot results in Fig 1D (page 1372, first paragraph on the right column) is corrected as follows: The depicted analyses of p‐p53, p‐p38, and GAPDH in Fig 1D (see also the corresponding Source Data file in the original article) were all analyzed from the same blot. A repeat of the p‐p53 Western blot was run on a separate gel using the same protein lysates and loading buffer (see Source data for Figure 1D below).
The Western blot for p21 in Fig 1D and the corresponding beta‐actin control were run in parallel with the same protein lysates and the same loading on 2 separate gels; the gel probed for beta‐actin was not shown in the original source data file of Fig 1D. The gel was also used to repeat the analysis of p‐p38 expression (see Source data for Figure 1D below). Results of p21 induction were confirmed by immunofluorescence analysis (see Fig 1E and Supplementary Fig S2H–K of the original manuscript).
The corrections do not affect the original conclusions presented. The authors apologize for the mistakes and any inconvenience they might have caused.”
Editorial Note
These changes were based on the results of an investigation by the Leibniz Association. All authors agree with this correction.




- Tao S, Tang D, Morita Y, Sperka T, Omrani O, Lechel A, Sakk V, Kraus J, Kestler HA, Kühl M, Rudolph KL. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 2015 Mar 4;34(5):624-40, Figure 1E und 1F. Correction 2.10.2017
“The authors state that it came to their attention that the figure legends of two figures were not sufficiently detailed in the original version of the manuscript.
The figure legend of Fig 1E and F should read:
A single representative FACS plot was chosen to illustrate (E) the gating of LGR5hi and LGR5lo populations within the population of LGR5+ (GFP‐positive) intestinal cells and (F) the gating of LGR5hi‐high, LGR5hi‐low, LGR5lo‐high, and LGR5lo‐low populations. The same FACS plot is displayed in (E) and (F).
The figure legend of Fig 6C should read:
Representative Western blots of cell lysates for the expression of phospho‐p53 and cleaved caspase‐3 (each n = 3; see Source Data for this figure). Samples from a single experiment were divided into identical portions and probed for phospho‐p53 or cleaved casp3. The expression control of beta‐actin was conducted on the second aliquot of the samples and was run in parallel on a separate gel.
The corrections do not affect the original conclusions presented. We apologize for the lack of detail and any inconvenience it may have caused.”
Editorial Note
These changes were based on the results of an investigation of the Leibniz Association. All authors agree with this correction.

- Hartmann K, Illing A, Leithäuser F, Baisantry A, Quintanilla-Martinez L, Rudolph KL. Gene dosage reductions of Trf1 and/or Tin2 induce telomere DNA damage and lymphoma formation in aging mice. Leukemia. 2016 Mar;30(3):749-53, Figure 1F. Corrigendum 10.11.2017
“Following the publication of this article the authors noted that the article was lacking the information on n-numbers for some of the panels of the figures.
The description of the results section is specified for the following information on n-numbers of mice: The western blots in Figure 1D and Supplementary Figure 1F were conducted as biological replicates (testis and thymus) using pools of mice of the indicated genotypes (n=6 mice per genotype for thymus, n=4 mice per genotype for testis). The quantification of the expression of telomere binding proteins was conducted on RNA level (Figure 1a and b; Supplementary Figure 1A–D, n=3 mice per genotype) and on protein level (Figure 1c; Supplementary Figure 1E, wild type: n=6 mice, Trf1+/−Tin2+/−: n=7 mice, Trf1+/− mice: n=4 mice, Tin2+/−: n=4 mice). In the latter analysis, one of the total seven wild-type mice was excluded due to an oversaturated GAPDH signal by statistical outlayer analysis (Grubbs’ test). The quantification of the telomeric signals of telomere binding proteins (Figure 1f and g; Supplementary Figure 1H) was carried out on a total of 37–43 nuclei per genotype from three independent cultures of mouse embryonic fibroblasts. The tumor-free survival curves (Figure 1j) and the pie chart on tumor formation (Figure 1k) were calculated from the identical number of mice as shown in Supplementary Figure 1J. For Figure 1k all mice from the three different knockout cohorts were combined.
The authors wish to apologize for any inconvenience caused.”
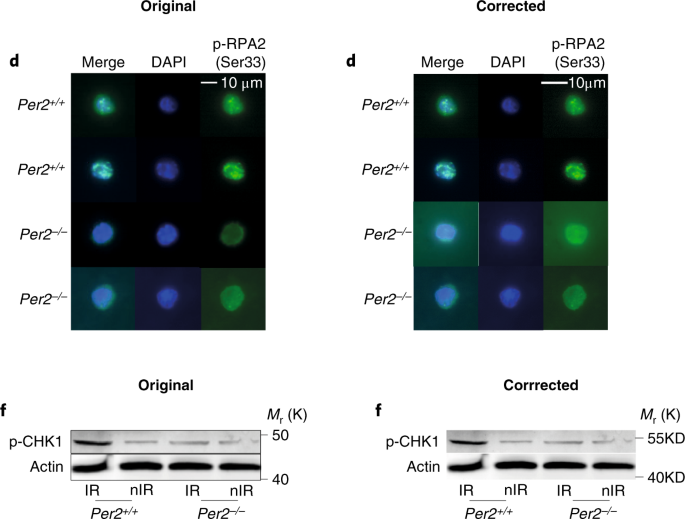
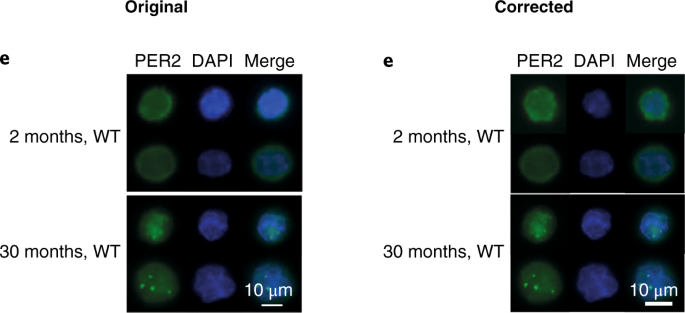
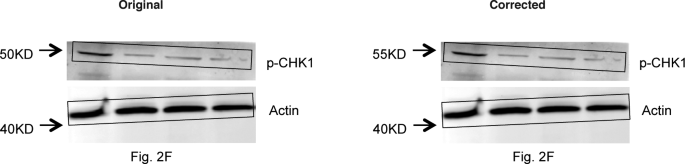
- Wang J, Morita Y, Han B, Niemann S, Löffler B, Rudolph KL. Per2 induction limits lymphoid-biased haematopoietic stem cells and lymphopoiesis in the context of DNA damage and ageing. Nature Cell Biology 2016;18:480-90, Figure Suppl. 6 Correction 26.02.2019
“In the version of this Article originally published, the authors mistakenly used the same images for the Fig. 2d Per–/– upper Merge, DAPI and p-RPA2 (Ser33), and Fig. 3e upper 2 months, WT Per2, DAPI and Merge panels. The correct images from these experiments, and their correctly sized scale bars, are shown below. In addition, the full dataset of representative images for Figs. 2d and 3e, and the numerical source data of the corresponding quantifications of Figs. 2c and 3f, have been uploaded to Figshare (https://doi.org/10.6084/m9.figshare.c.4353791.v1). In addition, the Methods section did not provide sufficient detail on how immunoblot experiments were conducted. Western blots were repeated three times using the antibodies stated in the Methods. PER2 blots were obtained using the PER2 antibody (Millipore, AB2202); except in the PER2 blots in the Repeat 1 experiments corresponding to Figs. 3g and 3k, where the PER2 antibody from BD Transduction (611138) was used. Repeats 1 and 2 were imaged using ECL and LI-COR methods, and Repeat 3 only used the LI-COR system. The images presented in the paper were from Repeat 3 and were stored electronically and as printouts, however the electronic versions were cropped as depicted in Supplementary Fig. 6. Repeat 1 images were stored electronically and Repeat 2 images only as printouts. In addition — with the exception of Fig. 2g (ATM), 3g (PER2, Repeat 1A), 3k (PER2, PUMA, p21, BAX, BCL2) and Supplementary Fig. 1a of Repeat 1, in which actin immunoblots were loading controls processed from the same membranes used to detect the target proteins — in all other cases the actin blots were sample processing controls that were run in parallel using the same lysates and loading concentrations that were used for the detection of the target proteins. The electronic images from Repeat 1 and the printout images of Repeats 2 and 3 have been uploaded to Figshare (https://doi.org/10.6084/m9.figshare.c.4353791.v1). Finally, there is an error in the protein size label of the p-CHK1 blot of Fig. 2f and the corresponding blot in Supplementary Fig. 6, which should read ’55KD’ instead of ’50KD’. The corrected panels are shown below.”
Update 11.01.2021
I noticed the above links to the Jena JAM 2020 conference were deleted, even the archived version I offer above was removed, impressive. Anyway, I didn’t miss anything, the meeting was postponed till 2021 due to COVID-19 (archived version here). New date will be announced soon!

There still was not a single retraction for Rudolph, but new corrections from January 2021: Tang et al JEM 2016 and Begus-Nahrmann et al JCI 2016 (“the described corrections do not change any of the conclusions of the article“). Oh, and FLI has now a “Good scientific practice” page where we learn:
“Good scientific practice (GSP) is the essential requirement for sustainable success, trust and long-term recognition in science. Accordingly, the staff of the FLI and its predecessor institute have practiced GSP for more than 25 years and contributed to the good national and international reputation of this Leibniz Institute.”
Neither Rudolph, not his research misconduct affair are ever mentioned. It’s not important anyway, Rudolph still receives grant funding from the DFG, including a new project from 2019.
Disclaimer: in my past as stem cell researcher, my work overlapped at some point with that of Rudolph in what can be considered as a scoop, after Rudolph was shown my unpublished results. His resulting paper Wang et al Cell 2012 is on the Leibniz list of investigated papers and also one of 3 papers flagged by DFG for research misconduct. I also once unsuccessfully applied for a faculty position in Jena where Rudolph was selection committee member. LS.

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism, however small it appears to you, will greatly help me with my legal costs.
€5.00


I already gave up going to certain meetings as these meetings are full of cheaters…not only one cheater but a congregation of them as they all protect each other
LikeLike
I think this is the larger problem that really needs identifying and addressing. The ‘club’ and the protections that is conferred to ‘members’ is unbelievable. I have experience and knowledge of multiple failings of Heads of Department that protect their PIs, despite absolutely despicable (fraudulent research, unethical practices, harassment (of all forms), bullying, etc.) behaviour. I have really lost faith in academic institutions as they are just not interested in promoting good practice and do all they can to protect one another.
LikeLike
I went to a Carlo Croce talk just before his shit hit the fan. In his talk, he showed a picture of his wall filled with masterpieces (and he used this word…he didn’t say “paintings”, he said “masterpieces”, with his thick Italian accent!). What a bloated (literally, and figuratively) wad this man is. That, in comparison to my crap salary (which may soon end), makes me realize what an incredibly unethical system I am a part of. There needs to be a Gini factor for scientists, which is right next to the i10-factor on google scholar, so I can avoid those talks as well if their factor is near 1.
LikeLike
Now when I review papers citing studies with known “problems”, I advice the authors to refrain from citing the studies I find questionable based on appropriate links to Pubpeer, RetractionWatch or For Better Science. Thus far, I received no push back from editors and authors from doing that.
LikeLike
He has his own audience. Germany anyway has to face the problem soon that its medical science community is dominated by MD professors who are no more than B-category researchers.
LikeLike
Talking about Mario Saad:
https://www.fcm.unicamp.br/fcm/relacoes-publicas/saladeimprensa/um-ensaio-clinico-sobre-fake-news-por-mario-saad
LikeLike
Thanks, I also incidentally found it last month. Comedy Gold, only not funny in Brasil.
LikeLiked by 1 person
Another note: Leonid, do you consider to check publications from the co-authors of Rudolph? You had posts here about WB loading controls. I was expecting you to recognize that his previous Jena-based coworkers have similar data irregularities in their own papers.
LikeLike
Mismatched loading controls are a tricky thing. Everyone knows of course the data is rigged, why else would one run loading control on a separate gel. But if you start to criticise, they scream back: how dare you not to trust our honesty!
LikeLike
There is also Photoshop artistry in those papers, not just mismatched loading controls. Moreover: study design errors, statistical miracles, incredibly low error bars. Who searches for them, will find them.
LikeLike
A brilliant suggestion. Never accept or support research misconduct. They seem strong but are weak. In a first step the researchers try to defend themselves, in a next step people around them defend themselves. When requesting open data a third stage occurs. If no open data is shown they should never have fundings again.
LikeLike
Pingback: The wizard men curing breast cancer – For Better Science
An obituary of Günther Niethammer at https://onlinelibrary.wiley.com/doi/pdf/10.1111/j.1474-919X.1975.tb04230.x does not refer to a paper by Günther Niethammer about observations of birds at and around Auschwitz in the period October 1940 – August 1941. See https://archive.org/stream/DieVogelweltVonAuschwitz#mode/1up for this paper (in German, with illustrations). Günther Niethammer was a long-term Editor-in-Chief of the Journal of Ornithology https://link.springer.com/journal/10336 See for backgrounds also https://www.amazon.com/Biologists-Age-Totalitarianism-Eugeniusz-Nowak/dp/1527510999
LikeLike
Dr Hans Kummerlöwe aka Dr Hans Kumerloeve was a close friend of the above mentioned Günther Niethammer. Kummerlöwe was born in 1903 and died in 1995. I can vaguely remember from those days to have read in an ornithological journal a ‘friendly’ obituary about his ornithological activities.
The research by Dr Eugeniusz Nowak (published in English in ‘Biologists in the Age of Totalitarianism’,
https://www.amazon.com/Biologists-Age-Totalitarianism-Eugeniusz-Nowak/dp/1527510999 , reveals a different picture.
“A Junker [landowner] from east of the Elbe (….) recalled his participation at the 8th International Ornithological Congress in 1934 in Oxford (…): ‘(….) not only wore the Party badge very conspicuously but also sat at the piano as a guest of the venerable Exeter College and played the Horst Wessel song in front of the open window, which could be heard far and wide.'(…)”
“May 1938: Participation in the 9th International Ornithological Congress in Rouen in France: Kummerlöwe came to the festive banquet with the Gold Metal of the NSDAP on the chest.”
“Kummerlöwe published his ‘scientific-political’ credo in two lengthy publications in 1939 and 1940. But incredibly the first one has been removed from most German libararies by unknown perpetrators, or pasted over; I received a copy for my research from the University Library in Copenhagen. To my amazement this text has also been partially pasted over in the Library of the Zoological Institute in Warzaw; in many libraries both are missing altogether! There are repeated references to Adolf Hitler or the Party, as well as terms like ‘protection of out blood’, ‘the racial sense of responsibility’, ‘the struggle for survival of our people’ and alot more of the same.”
“Professor Jochen Martens of Mainz wrote to me (…) (20.7.2000): (…..) “I was, and am, of the opinion that Kummerlöwe removed them himself from the libraries after the war, thus providing himself with a ‘clean slate’.””
“The financial support that he received during the postwar period as a private researcher was extremely generous. (…) I turned to the DFG in Bonn with a request for more exact details. I was allowed to read the printed annual reports (….) as early 1949 (…) he received money to process his observations on birds on the island of Amrum. A total of at least 23 advances of money were marked to Kummerlöwe in the annual reports, 7 times for materials, 13 times for travel, and 3 times for printing. The travel grant allowed him to travel to Belgium, France (3 times), to England, to Turkey (7 times), to Syria, and to Lebanon as well as Morocco (2 times). (…) He recieved his last material grant in 1987, that is as an 84-year-old! Did the DFG have more money at that time than today or was this applicant just ‘luckier’ than the others?”
“Scientists of today can only dream of such finances! Even I dreamed of support from the DFG for my biographical research, making for instance a detailed application for the purpose of personal reseach on Kummerlöwe’s work in Vienna. The replies I received (9. and 17.1.2001) requested me to understand that the rejection was not only because of the ‘limited funds’ of the DFG, but also because of the fear that I would not be able to apply the ‘modern criteria of oral history’ and that I would not be able to cope with the ‘difficult records’ in the archives.”
See for backgrounds also http://muscicapa.blogspot.com/2017/07/shocking-tales-from-ornithology.html
LikeLike
Pingback: Amato Giaccia: too big to fall – For Better Science
Pingback: Jan van Deursen left Mayo Clinic – For Better Science
https://www.jci.org/articles/view/143687 Conversations with Giants in Medicine 10.1172/JCI143687 A conversation with C. Ronald Kahn
Ushma S. Neill First published October 1, 2020
http://retractiondatabase.org/RetractionSearch.aspx#?auth%3dKahn%252c%2bCarl%2bRonald A retraction (senior author), expression of concern (senior author) and a correction in the very same journal
LikeLike
2021 erratum for:
FASEB J . 2014 Oct;28(10):4408-19. doi: 10.1096/fj.14-253971. Epub 2014 Jul 8.
Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications
Cecile Vernochet 1, Federico Damilano 2, Arnaud Mourier 3, Olivier Bezy 1, Marcelo A Mori 1, Graham Smyth 1, Anthony Rosenzweig 2, Nils-Göran Larsson 3, C Ronald Kahn 4
Affiliations
1 Section on Integrative Physiology and Metabolism, Joslin Diabetes Center, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA;
2 Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA; Cardiovascular Division, Department of Medicine, Beth Israel Deaconess Medical Center, Boston, Massachusetts, USA; and.
3 Department of Mitochondrial Biology, Max Planck Institute for Biology of Ageing, Cologne, Germany.
4 Section on Integrative Physiology and Metabolism, Joslin Diabetes Center, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA; c.ronald.kahn@joslin.harvard.edu
https://pubpeer.com/publications/57CEB9AB62EF9CEAF3678F73DA0A51
2021 erratum. https://faseb.onlinelibrary.wiley.com/doi/10.1096/fsb2.21633
The authors report that in Figure 1B, the western blots labeled inguinal white adipose tissue (iWAT) tubulin and brown adipose tissue (BAT) mitochondrial transcription factor A (TFAM) contain duplicated images. Note, in this series of experiments, tubulin served as a loading control for TFAM and was used for quantitation. After careful re‐examination of the original data, it appears that this was a duplication; while the BAT TFAM blot is correct, a copy of the same blot was incorrectly inserted in the position of the iWAT tubulin blot. After identification of the correct blot and revision of the figure, recalculation of the quantitation shown in the right panel of Figure 1B revealed no change from the original, indicating that the mistake was solely an image duplication error. Thus, this error did not affect the validity of the research or the conclusions drawn in the paper. The authors thank the reader who brought this mistake to their attention and apologize for any confusion that this error may have caused.
LikeLike
Someone apparently wants a Nobel prize,,,,,
https://www.jci.org/articles/view/133156/figure/5
LikeLike
Lots of famous person ass-kissing and back-slapping going on here. Pretty disgusting.
https://www.jci.org/articles/view/133156/sd/5
LikeLike
Ron Kahn, like Carlo Croce, likes to buy art with his prize money. So we have some old-fart MD advisor sitting on his ass off the hard work of his honest post-docs and the ones that fabricate data, buying art with his money.
https://www.bizjournals.com/boston/blog/bioflash/2015/11/joslin-doctor-given-nobel-prize-in-diabetes.html
No wonder people storm the capitol or execute the royal family.
LikeLike