France is a science superpower, the orders to maintain the highest standards of research misconduct arrive from the very top: the Academie des Sciences and CNRS leadership, even the Minister for Research is a biomedical researcher with her own problems. Frederique Vidal, who is a former university rector, recently had a retraction, after her ministry twice used a paywalled newspaper to threaten to sue me for disclosing data irregularities in her works. Vidal’s own University of Nice has other issues, as evidenced in the PubPeer-listed papers of Nice professors Bernard Rossi, Heidy Schmid-Antomarchi, Annie Schmid-Alliana, Jean-Francois Peyron and Dorota Czerucka.
But this is actually not about Vidal, or University of Nice, or the previous affairs at CNRS I wrote about, or even this new project by data integrity sleuth Clare Francis, about the Lyon University stem cell biologist Michelina Plateroti. Seriously, I cannot keep up, so instead a story of the cancer researcher Patrick Legembre, a young group leader at University of Rennes, whose lab is funded by INSERM.
When evidence of data manipulation appeared on PubPeer, Legembre bravely stepped in and explained everything. To cut a long story short: none of the conclusions were affected. It is apparently a great French tradition where the scientists under suspicion of research misconduct function as their own investigators, as it happened in the case of former CNRS chief biologist Catherine Jessus, and even the disgraced plant scientist Olivier Voinnet tries to established himself as his own chief investigator.
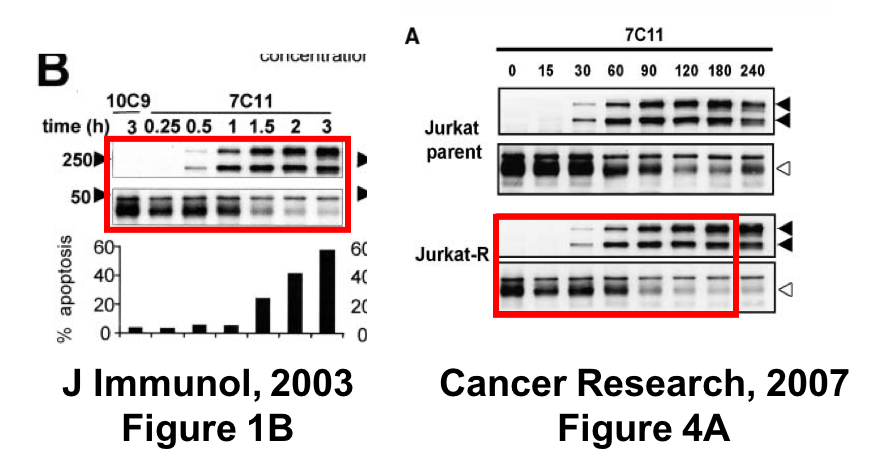
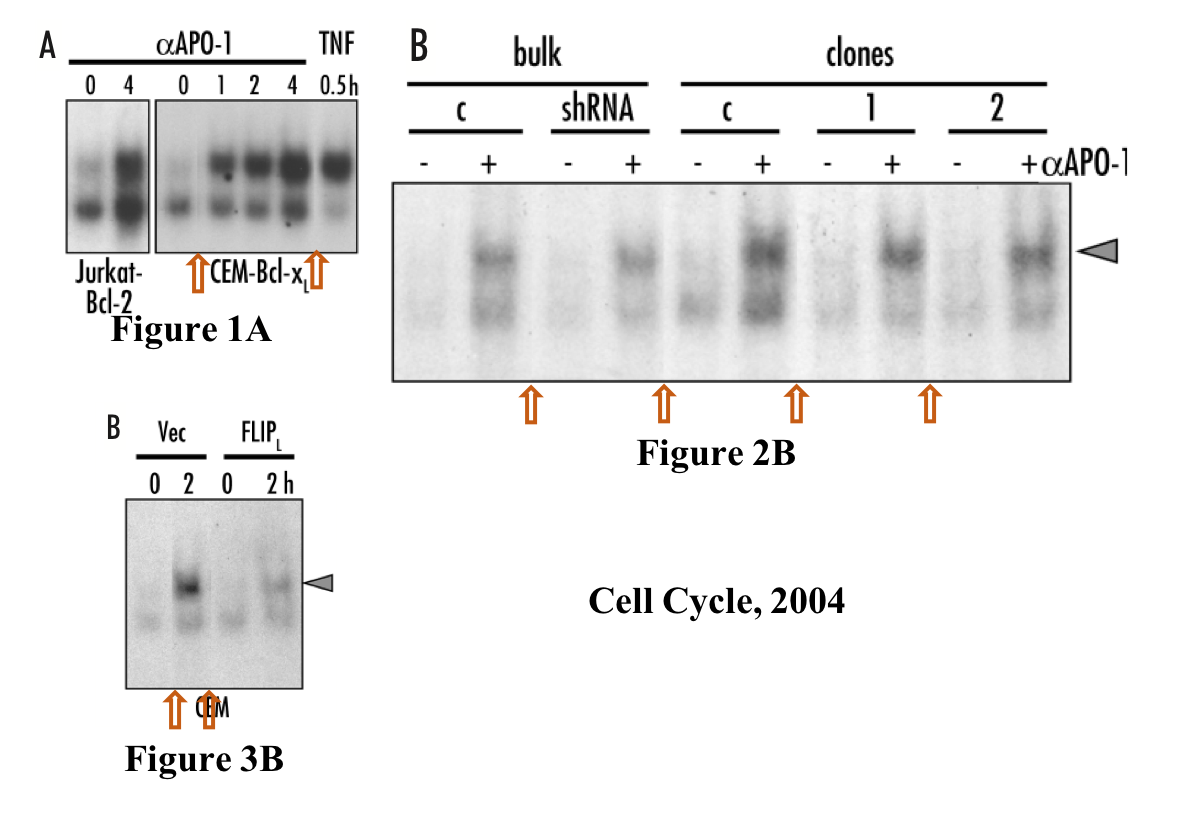
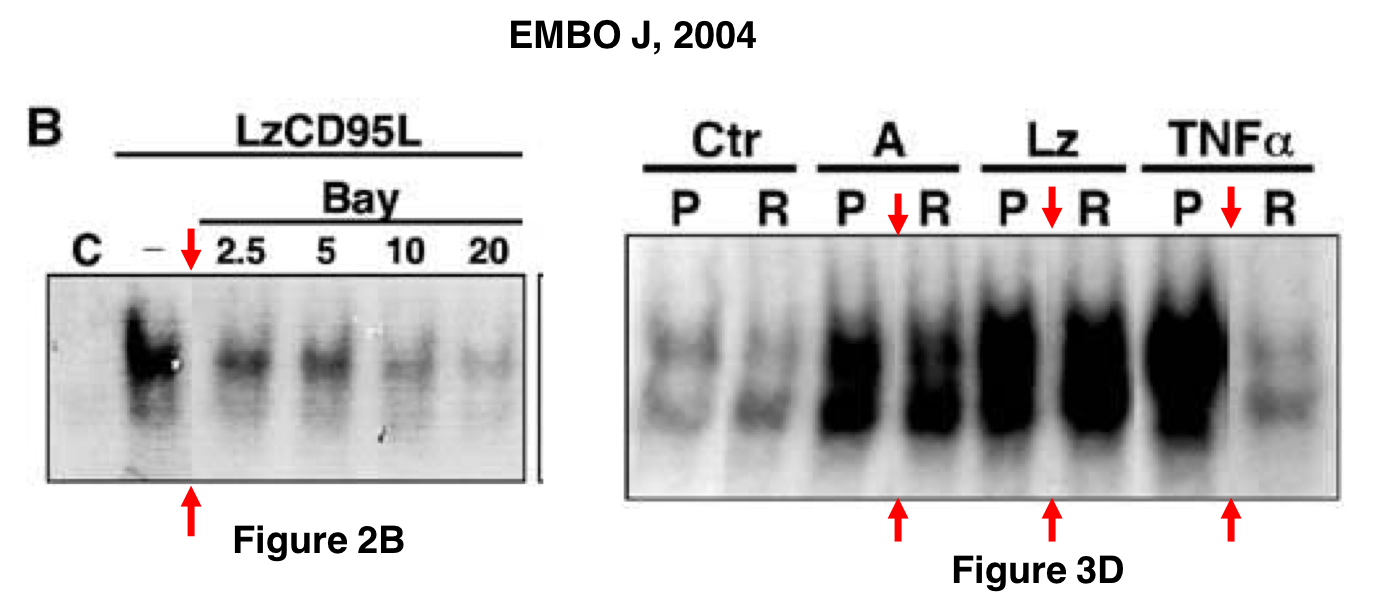
This was found in the paper Beteneau et al Cancer Research 2007, and shared on PubPeer:
The last author Legembre shared the original blots and explained:
“These original gels confirmed that Jurkat cell (parental cells) data have been reused and unfortunately, have been switched for Jurkat-R cells in the Cancer Research manuscript. The pattern of CD95-SDS stable microaggregate formation is similar between the two cells and this could explain this inadvertent inversion. This reuse (J immunol, 2003) of the parental cell and its inversion is highly unfortunate but does not change the message…”
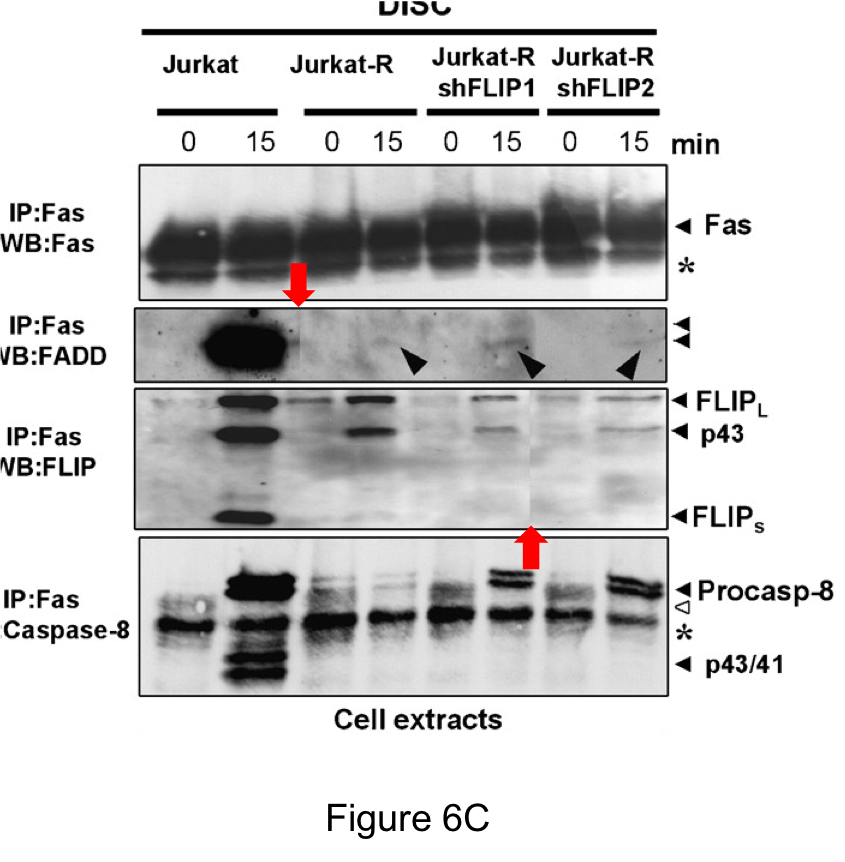
The paper also contained a spliced image, making it obvious that the FAS loading control was on a different gel from the other gels (at least 2 more):
Also here, Legembre tried to assuage, worried about the journal’s possible scepticism:
“there is one cut present for the c-FLIP and FADD (cut between clones Jurkat-R shFLIP1 and 2) immunoblots because clone 2 and 3 were shifted during the gel loading in these gels. Although these inadvertent mistakes do not affect the scientific message of the presented data, we are sorry for these errors. Cancer research has been contacted for a corrigendum.“
The accidental data reuse in Legembre lab seems epidemic. This happened in Fouque et al J Medicinal Chemistry 2015, as reported on PubPeer in September 2019. The publisher ACS immediately issued a correction.
Fig. 6g: Same cell death curves for Jurkat and CEM for DHM25 condition
Sup. Fig.1, right panels: Akt-p and total Akt look similar”
Legembre explained:
“Indeed, an error occurred with the two MTT assays, and T47-D was, in fact, a copy-past of the MDA-MB-231 MTT. T47-D MTT has been replaced, fitting now the IC50 depicted in Figure 2 B (see attached figures). In figure 6D, the cytotoxic effect of DHM25 on CEM cells monitored by MTT assay was identical to the effect measured in Jurkat cells. The mistake stems from CEM data and has been corrected. The immunoblots in supporting Fig.1 were double-checked and they are not coming from an unfortunate copy and paste. The similarity comes from the process of antibody labeling and stripping (P-Akt blot, stripping and Akt reblotting).”
It can indeed happen one accidental recycles a dataset where all labelling is same except the cell line used, as in Figure 2a. Less clear is how one accidentally re-uses only one control between two datasets of two different cell types. And maybe ACS chemistry experts do not know that total and phosphorylated protein western blots should look similar, but not identical.
More innocent mistakes happened in Malleter et al Cancer Research 2013, and reported on PubPeer in September 2019.
Last author Legembre was ready to fix the problem:
“The CD95 control (lysates) for CD95 immunoprecipitations in MDA-MB-468 cells (Figure 5D) was missing and instead, two c-yes loading controls (src kinase) were depicted. We modified the figure by adding the CD95 control for this experiment (see attached figures). Boyden chamber images of the MDA-MB-231 and MDA-MB-468 TNBC cells were identical in Supporting Figures 5C and 5H. The mistake came from pictures shown for the MDA-MB-468 cell line. Therefore, we replaced these pictures by the correct images (see attached figures). These modified figures have been sent to the editor of Cancer Research for correction.”
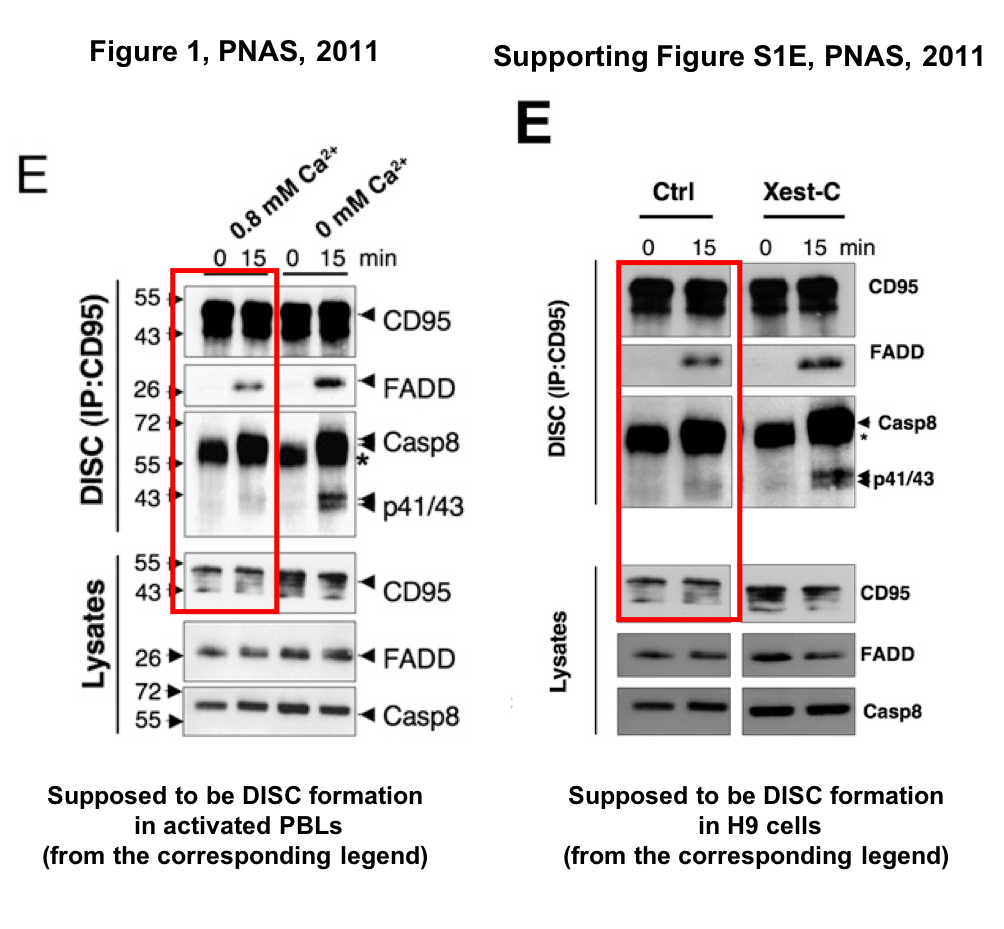
A similar unfortunate switch of cell lines and results happened in Khadra et al PNAS 2011, as discussed on PubPeer. This paper was “Contributed by Michael D. Cahalan”, a coauthor who is member of National Academy of Sciences USA. Meaning, the manuscript was published in PNAS without a proper peer review, an insider route routinely abused to publish poor quality science, bunk or even outright data fabrications.
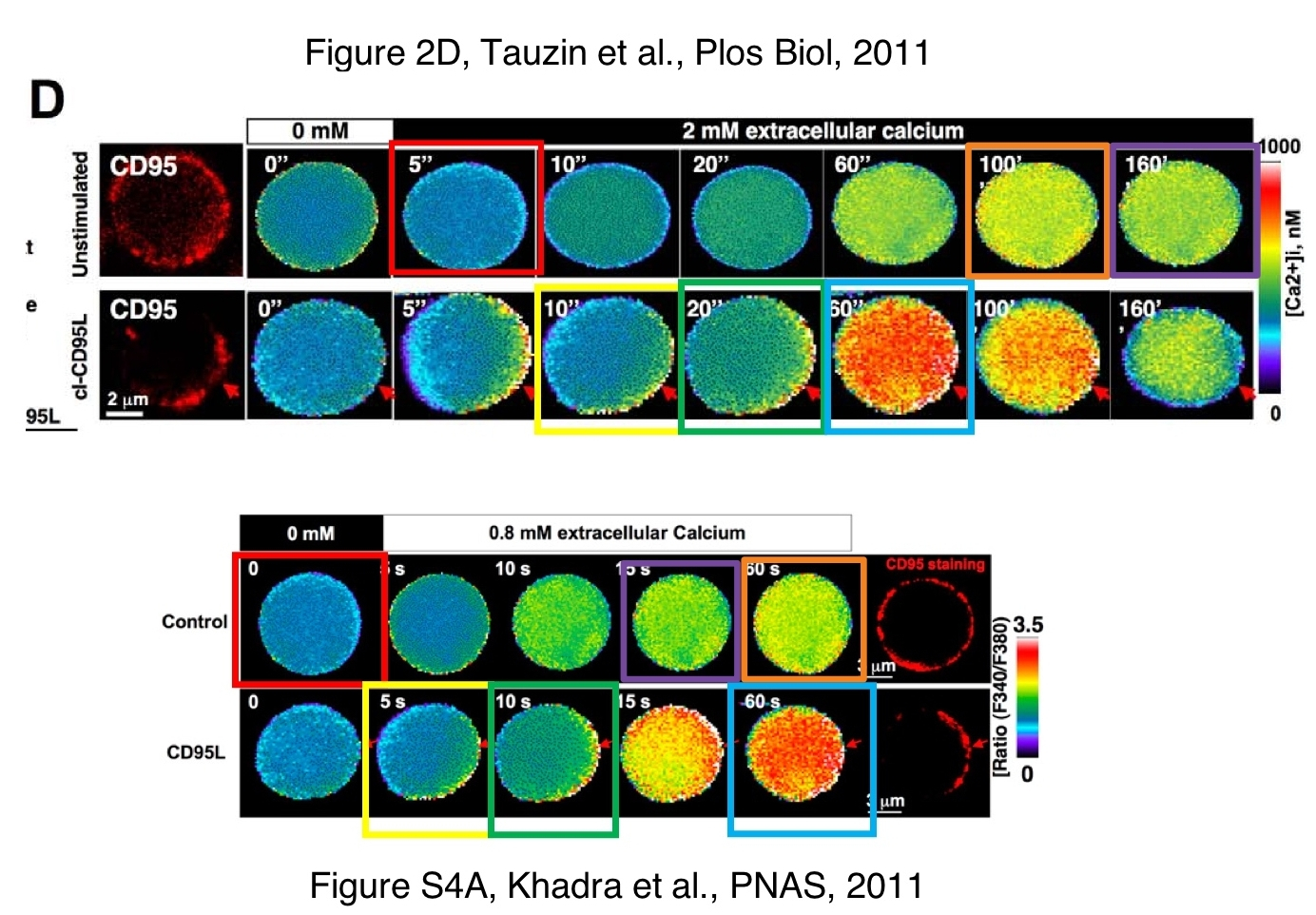
There were also reused graphs as well as beautifully coloured image re-assemblies from the paper Tauzin et al PLOS Biology 2011, where Legembre also happens to be last author:
That PLOS Biology paper had even more issues, maybe it was too fast of the publisher PLOS to correct it just on 8 October 2019 for what appears to a casual reader as a mere typo. The impactful study namely served 5 years later as template for the paper, Monet et al, Scientific Reports 2016 and was criticised on PubPeer in September 2019:
One can poke fun at Nature’s garbage bin Scientific Reports and their quality control, but it is still puzzling why PLOS Biology had no issue with empty phospho-AKT western blots lacking a positive control. If a PhD student brings this to a PI, the student gets sent back to the lab, to repeat the gel with a loading control. If an INSERM PI brings same useless gel to an elite journal, he gets congratulated on a paper accepted. But then again, Nature Publishing Group journal Oncogene accepted from Legembre an utterly blank picture (which never ever looks like a real gel) as stand-in for vimentin, in Edmond et al 2015. But then again, Oncogene is a very special journal, what’s with its Editor-in-Chief Justin Stebbing.
In any case, the penultimate author Legembre explained about his Scientific Reports recycling:
“In the PLoS Biology paper, we represented data obtained with PS120-NHE1 cells reconstituted with WT or (1-175) CD95 constructs and inadvertently, we indicated PS120 instead of PS120-NHE1 cells in “material and methods” Therefore, we contacted editors of PLoS Biology to replace “the hamster fibroblastic cell line PS120” by “the hamster fibroblastic cell line PS120-NHE1 designated PS120 in the manuscript” in Material and Methods session of this paper.“
Hence, what PLOS Biology so eagerly corrected, was not a mere typo, but a key feature of the cell line as well as the rather relevant point of the presence vs absence of NHE1 gene product. A scientific result was retrospectively significantly changed, to accommodate an image duplication in Scientific Reports. This is truly clever of Legembre, and I salute him and PLOS Biology for that.
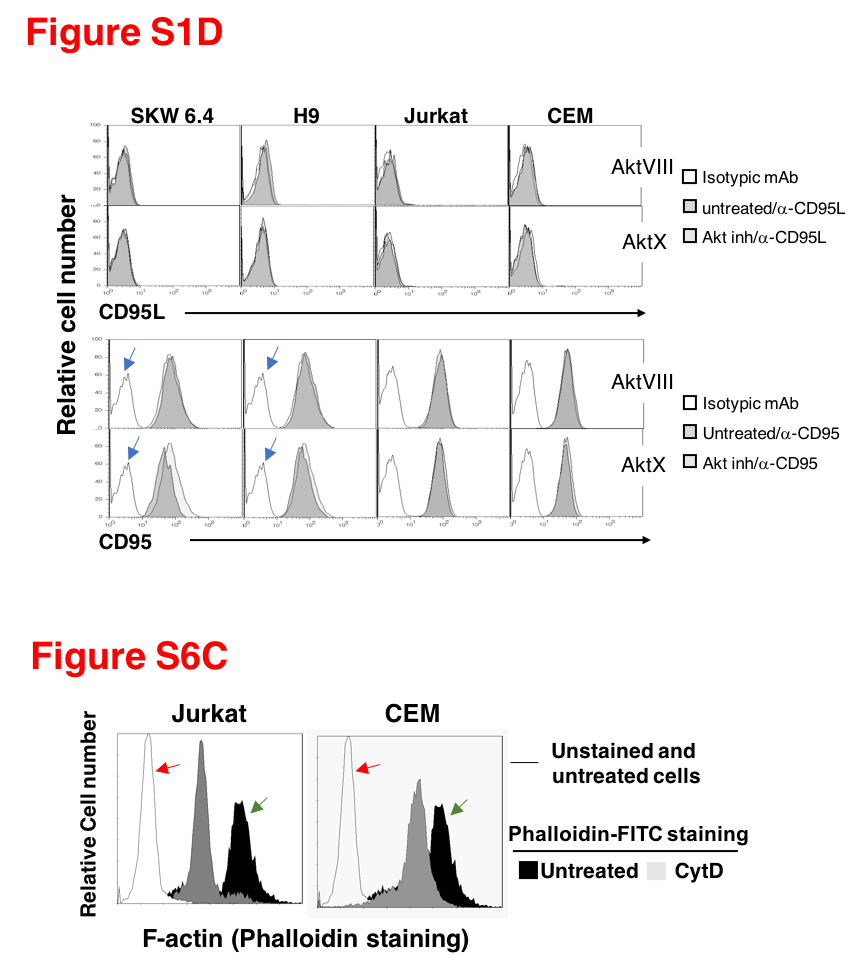
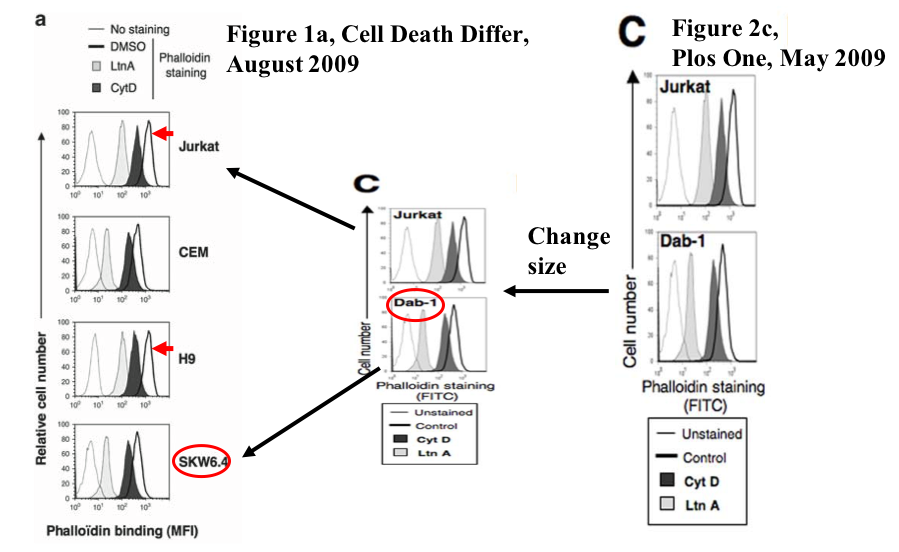
Some other innocent mistakes are as yet of uncommented by Legembre. It seems, in his lab no dataset is safe from accidental duplication: western blots, spectrometer multiwell assays, even flow cytometry, like in Pizon et al Eur J Immunology 2011. Again, different cell lines share same FACS profiles, and what makes it peculiar: only some sample data in the combined dataset panel was reused.
A similar thing happened in Edmond et al PLOS One 2012, as recorded on PubPeer. And before that, flow cytometry data was exchanged between Chaigne-Delalande et al Cell Death Diff 2009 and Guidicelli et al PLOS One 2009, the former paper also copy-pasted a CD95 western blot for two different cell lines, as discussed on PubPeer, and yet not commented by the last author and chief investigator of his own papers, Patrick Legembre.
Maybe the Legembre lab is constantly off their faces on cheap cider, would that explain constant mix-up of published data? But then again, are we still sure the conclusions are not affected?
To be fair, Legembre himself never had a strong enough grasp on the concept of data integrity. This paper Legembre et al Cell Cycle 2004, from his postdoc period with Marcus Peter at the University of Chicago , USA, visualises it.
Already in 2004, when gel splicing in Photoshop was allegedly perfectly accepted, Legembre obviously knew that this practice must be indicated with vertical lines or empty space, as he actually did in Figure 1A. But for some reason, he chose not to indicate the other 8 lane splicing events in the same paper, including that same Figure 1A. Legembre was busy splicing gels also in these Peter lab papers Legembre et al J Biol Chem 2004 and Barnhart et al EMBO J 2004:
Same practice was applied back in France, when Legembre became junior PI in the lab of Jean-Francois Moreau at University of Bordeaux, in Legembre et al Mol Cell Biol 2005.
With so much growing PubPeer evidence, Legembre was only able to comment about his PNAS paper:
“We are gathering data to present a corrected version to PNAS editors. This takes some time due to the fact that these data came from different laboratories.”
Translation: in case the conclusions are affected after all, it was someone else who did it.

Update 17.10.2019
Two independent sources informed me that Patrick Legembre was accused of bullying and disparaging behaviour towards female subordinates, e.g. he allegedly introduced a female senior lab member to a colleague as “a lazy ass”. INSERM apparently lost in the administrative court when trying to have Legembre’s tenure revoked. He is now reintegrating. I will share more information when I receive it.
Update 18.10.2019
I contacted the INSERM press office with the information of the above update. I now received this reply from the head of press office, Priscille Riviere:
- “Monsieur Legembre est chercheur Inserm / Mr Legembre is researcher at Inserm
- Une procédure est toujours en cours et pour cette raison l’Inserm ne la commentera pas /Proceedings are still ongoing. For this reason, Inserm will not comment
- Notre délégation à l’intégrité scientifique est à la disposition de toute personne qui souhaite la saisir pour un doute ou une question sur des travaux scientifiques / Our Scientific Integrity Office can be seized regarding any doubts or questions on scientific researchs, by anyone wishing to do so.“
I followed the invitation and lodged an official notification of suspected research misconduct with Ghislaine Filliatreau, Inserm Scientific Integrity Officer
Update 2.04.2020
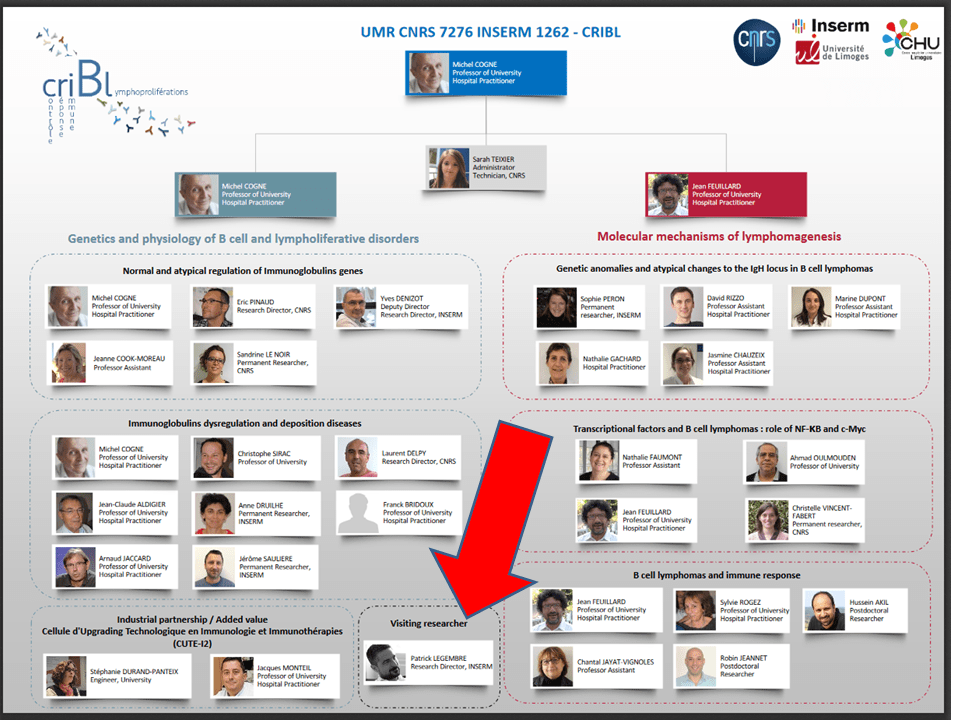
Legembre was apparently transferred from Rennes to a CNRS-INSERM institute in Limoges, where he is listed as “visiting researcher”. See this organigram from March 2020:

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism, however small it appears to you, will greatly help me with my legal costs.
5.00 €





“Errare humanum est, perseverare autem diabolicum” – Lucius Annaeus Seneca.
LikeLike
Pingback: No Time Toulouse! – For Better Science
“project by data integrity sleuth Clare Francis, about the Lyon University stem cell biologist Michelina Plateroti.”
2020 retraction.
PLoS One. 2012;7(4):e34162. doi: 10.1371/journal.pone.0034162. Epub 2012 Apr 3.
Multi-level interactions between the nuclear receptor TRα1 and the WNT effectors β-catenin/Tcf4 in the intestinal epithelium.
Sirakov M1, Skah S, Lone IN, Nadjar J, Angelov D, Plateroti M.
Author information
1
Institut de Génomique Fonctionnelle de Lyon, Lyon, France.
2020 retraction notice.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0228994
Following the publication and correction of this article [1, 2], additional concerns have been raised regarding Fig 2 and Fig 5:
In Fig 2C, lane 2 and 3 in the activated β-catenin panel, representing the T3 and the T3-Wnt3a samples, appear similar.
In Fig 2C, lane 1 and 3 in the Actin panel, representing the Ctrl and the T3-Wnt3a samples, appear similar.
In Fig 2C, vertical discontinuities in background have been detected between lane 3 and lane 4 of the Activated β-catenin panel, and between lane 1 and lane 2 in the Actin panel.
In Fig 5, the T3- and T3+ results for the WB:TRα1 Inputs panel appear similar.
In Fig 5, lanes 3, 4 (TRα1 IP) appear similar to lanes 5, 6 (β-catenin IP) in the WB:β-catenin panel.
In Fig 5, the T3- and T3+ results for the WB:β-catenin Inputs panel appear similar.
In Fig 5, irregularities have been detected in background granulation just below the Inputs (+) band of the WB: β-catenin panel.
The authors have indicated that the underlying primary data for the figures above are no longer available.
In light of the concerns affecting multiple figure panels that question the integrity of these data, the PLOS ONE Editors retract this article.
MS, SS, INL, JN, DA did not respond. MP did not agree with retraction.
References
1.Sirakov M, Skah S, Lone IN, Nadjar J, Angelov D, Plateroti M (2012) Multi-Level Interactions between the Nuclear Receptor TRα1 and the WNT Effectors β-Catenin/Tcf4 in the Intestinal Epithelium. PLoS ONE 7(4): e34162. https://doi.org/10.1371/journal.pone.0034162
pmid:22509275
2.The PLOS ONE Staff (2014) Correction: Multi-Level Interactions between the Nuclear Receptor TRα1 and the WNT Effectors β-Catenin/Tcf4 in the Intestinal Epithelium. PLoS ONE 9(3): e93418. https://doi.org/10.1371/journal.pone.0093418
LikeLike
2020 editor’s note for:
Cancer Res. 2007 Jan 1;67(1):108-15.
Dominant-negative Fas mutation is reversed by down-expression of c-FLIP.
Bénéteau M1, Daburon S, Moreau JF, Taupin JL, Legembre P.
Author information
1
Laboratoire Composantes Innées de la Réponse Immunitaire et Différenciation, Centre National de la Recherche Scientifique UMR 5164, University of Bordeaux 2, 146 rue Léo Saignat, 33076 Bordeaux, France.
2020 Editor’s note.
https://cancerres.aacrjournals.org/content/80/2/363
The editors are publishing this note to alert readers to concerns about this article (1). The same Western blot analysis image was used for Jurkat-R cells in Fig. 4A and for Jurkat parental cells in Fig. 1B of another article previously published by the authors (2). In addition, lane 1 of Fig. 4A in this article is labeled as a 0 timepoint with 7C11 treatment, and in Fig. 1B in ref. 2 it is a 3 h timepoint with 10C9 treatment.
References
1.↵Bénéteau M, Daburon S, Moreau JF, Taupin JL, Legembre P. Dominant-negative Fas mutation is reversed by down-expression of c-FLIP. Cancer Res 2007;67:108–15.
2.↵Legembre P, Beneteau M, Daburon S, Moreau JF, Taupin JL. Cutting edge: SDS-stable Fas microaggregates: an early event of Fas activation occurring with agonistic anti-Fas antibody but not with Fas ligand. J Immunol 2003;171:5659–62.
LikeLike
2020 correction for:
Cancer Res. 2013 Nov 15;73(22):6711-21. doi: 10.1158/0008-5472.CAN-13-1794. Epub 2013 Sep 26.
CD95L cell surface cleavage triggers a prometastatic signaling pathway in triple-negative breast cancer.
Malleter M1, Tauzin S, Bessede A, Castellano R, Goubard A, Godey F, Levêque J, Jézéquel P, Campion L, Campone M, Ducret T, MacGrogan G, Debure L, Collette Y, Vacher P, Legembre P.
Author information
1
Authors’ Affiliations: Inserm U1085-IRSET, Equipe Labellisée Ligue Contre Le Cancer; Université de Rennes-1; Centre Eugène Marquis, rue bataille Flandres Dunkerque, Rennes; ImmuSmol, 15 Rue Amiral Prouhet, Pessac; Inserm, U1068; Institut Paoli-Calmettes; Aix-Marseille Université, CNRS, UMR 7258, Marseille; ICO-René Gauducheau, Bd J. Monod, Saint-Herblain; Université de Bordeaux; Inserm U1045, Université Bordeaux Segalen; and Inserm U916, Institut Bergonié, Bordeaux, France.
2020 correction.
https://cancerres.aacrjournals.org/content/80/3/639
In the original version of this article (1), the Western blot image used in Fig. 5D to represent CD95 lysate was inadvertently duplicated from the image of c-yes lysate. The image has been replaced with the intended image for CD95 lysate. In addition, in Supplementary Fig. S5H MDA-MB-468 cells, the cell migration images for control and cl-CD95L in the untreated group were inadvertently duplicated from the control group of Supplementary Fig. S5C MDA-MB-231 cells. The images have been replaced with the intended images for MDA-MB-468 cells. These corrections are reflected in the latest online HTML and PDF versions of the article. The authors regret these errors.
Reference
1.↵Malleter M, Tauzin S, Bessede A, Castellano R, Goubard A, Godey F, et al. CD95L cell surface cleavage triggers a prometastatic signaling pathway in triple-negative breast cancer. Cancer Res 2013;73:6711–21.
LikeLike
2019 correction for:
J Med Chem. 2015 Aug 27;58(16):6559-73. doi: 10.1021/acs.jmedchem.5b00991. Epub 2015 Aug 14.
A Novel Covalent mTOR Inhibitor, DHM25, Shows in Vivo Antitumor Activity against Triple-Negative Breast Cancer Cells.
Fouqué A1,2,3, Delalande O1,4, Jean M1,5, Castellano R6, Josselin E6, Malleter M1, Shoji KF1,7, Hung MD8, Rampanarivo H1,2,3,4, Collette Y6, van de Weghe P1,5, Legembre P1,2,3.
Author information
1
Université de Rennes-1 , 2 avenue du Prof. Leon Bernard, 35043 Rennes, France.
2
CLCC Eugène Marquis, INSERM ER440 Oncogenesis, Stress & Signaling, rue bataille Flandres Dunkerque, 35042 Rennes, France.
3
Equipe Labellisée Ligue Contre le Cancer, rue bataille Flandres Dunkerque, Rennes, France.
4
IGDR, CNRS UMR 6290, UFR des Sciences Biologiques et Pharmaceutiques, 2 Av. du Professeur Léon Bernard, 35043 Rennes, France.
5
UMR6226, CNRS, Institut des Sciences Chimiques de Rennes, Equipe Produits Naturels, Synthèses, Chimie Médicinale, UFR des Sciences Biologiques et Pharmaceutiques, 2 Av. du Professeur Léon Bernard, 35043 Rennes, France.
6
CRCM, INSERM, U1068; Institut Paoli-Calmettes; Aix-Marseille Université; CNRS, UMR 7258, Marseille, France.
7
INSERM, U1085, 2 avenue du Prof. Leon Bernard, 35043 Rennes, France.
8
Faculty of Chemistry, National Vietnam University , 19 Le Thanh Tong, Hanoi, Vietnam.
2019 correction.
https://pubs.acs.org/doi/10.1021/acs.jmedchem.9b01578
Page 6563. In Figure 2a, the plots for T47-D cells are incorrect. The correct Figure 2 is shown below, with the corrected area highlighted in red.
Page 6568. In Figure 6d, the cytotoxic effect of DHM25 on CEM cells monitored by MTT assay was identical to the effect measured in Jurkat cells. The mistake stemmed from CEM data. The correct Figure 6 is shown below, with the corrected area highlighted in red.
LikeLike
“Legembre was apparently transferred from Rennes to a CNRS-INSERM institute in Limoges”.
While in town he could visit these people and compare notes:
Clin Cancer Res. 2010 Oct 1;16(19):4711-21. doi: 10.1158/1078-0432.CCR-10-0470. Epub 2010 Sep 21.
Involvement of human herpesvirus-6 variant B in classic Hodgkin’s lymphoma via DR7 oncoprotein.
Lacroix A1, Collot-Teixeira S, Mardivirin L, Jaccard A, Petit B, Piguet C, Sturtz F, Preux PM, Bordessoule D, Ranger-Rogez S.
Author information
1
Centre Hospitalier Universitaire Dupuytren, Limoges, France.
Figure 5B. Much more similar than you would expect.
https://pubpeer.com/publications/B598A03663AAA17A6D3B71961F14FC
LikeLike
I understand that he has given the right Figures, wich doesn’t change any conclusion…
What about you ? If you come in town, what about comparing your work and his ? publication, IF etc etc…
Even if u’re right about those errors, i’m curious about how much you’re better than he is. Too curious ?
LikeLike
Pingback: For Better Science – Patrick Legembre
Pingback: mTOR: conclusions not affected? – For Better Science
Pingback: The Ballad of Claudio Hetz – For Better Science