Spanish science is in a disastrous state. The nation’s academia is infested with nepotism, political and religious networks, and massive, multi-generational research fraud. Honest scientists leave the country, those who remain are scared into silence. Fraudsters not only are not investigated, they even get their critics sanctioned for misconduct, fired, put on court trial, and make them apologise and beg for mercy in public.
We saw all this with the Carlos Lopez-Otin affair, the cruel travesty playing out in national media. But because nobody else does it, we at For Better Science continue exposing Spanish research fraud.
Welcome to a new post about dishonest plant scientists, by the pseudonymous PubPeer sleuth Aneurus Inconstans. It’s all about fake yeast assays.
Iberian yeast masters do it classic with a flip
By Aneurus Inconstans
Yeast are not plants, but fungi, yet they are an integral tool of plant science, which is particularly creatively applied in Spain.
Yeast-based techniques are used to verify protein-protein interaction, to test the functionality of recombinant proteins in yeast mutants, etc, and somehow many Spanish plant scientists love to play around with yeast strips beyond what is commonly legit. Nobody knows why. Maybe it’s part of academic culture?
You may remember the case of Pedro L. Rodriguez and his lab members at the Instituto De Biología Molecular Y Celular De Plantas (IBMCP) in Valencia, where yeast colonies were flipped, digitally modified and recycled to present different things in tens of papers over a period of fifteen years. The yeast problems weren’t the only ones though, there was a lot more. If you haven’t done so yet, I suggest you read Leonid’s articles (here and here) about that real-life comedy, which is a perfect blend of laughs and tears.
The following story takes place again on the Iberian Peninsula, this time in the sunny Sevilla. The peculiarity of this case does not lie in the volume of the articles discussed, rather on the psychological traits of the character involved.

José Román Pérez-Castiñeira, associate professor at University of Sevilla and author of the book “Chemistry and Biochemistry of Food“, is one of those scientists who seem to consider data manipulation as an acceptable side effect of their research activities, and he also likes to explain to you why it is acceptable, and he does it in great detail.
The long and lively discussion with Dr. Pérez-Castiñeira in the PubPeer thread of Drake et al. 2010 has it all. The article investigates the expression level and the subcellular localization of chimaeric membrane-bound pyrophosphatase H+-PPases from bacteria and eukaryotes in the yeast conditional mutant YPC3, which is lacking its essential cytosolic pyrophosphatase Ipp1p and is unable to grow on glucose. Chimaeric H+-PPases (or exogenous IPP1) expression would rescue YPC3 growth on glucose media. Take a look at the issues:
Drake R, Serrano A, Pérez-Castiñeira JR. N-terminal chimaeras with signal sequences enhance the functional expression and alter the subcellular localization of heterologous membrane-bound inorganic pyrophosphatases in yeast Biochem. J. (2010) doi: 10.1042/BJ20091491

the yeast strain utilized for its generation.
Figure 2A: Drop tests performed with YPC3 cells transformed with the genes coding for the cytosolic inorganic pyrophosphatase of S. cerevisiae (IPP1) and Trypanosoma cruzi H+-pyrophosphatase (TcVP) inserted in the cloning site of plasmid pRS699b. Negative controls are YPC3 cells transformed with plasmid pRS699b.
Pérez-Castiñeira replied on the problems in Figure 1C and on those in central panels of 2A (blue and red boxes) in a long and confused comment, discussing the characteristic of the strains used and the overall meaning of Figure 1. He also cited a previous paper by him that has its own serious issues, and which I’ll discuss later (hyperlink mine – A.):
“Cells are not that different, W303 is the parental strain of YPC3, the only change between both of them is the insertion of the GAL1 promoter controlling the expression of IPP1 in the latter. In my view, there is no surprise in these pictures because, although W303 (and most yeast strains) grows faster in glucose than in galactose we decided to wait as long as necessary in order to make sure that YPC3 had no growth whatsoever in glucose. […] Moreover, we had previously generated a mutant with the same phenotype as YPC3, albeit using a different experimental approach (Perez-Castiñeira et al., 2002). The former had the tendency to produce revertants after a few days, therefore, it was important for us to check that the insertion introduced in YPC3 was perfectly stable. For this reason, we incubated the plates…“


Nobody understands how this answer could possibly address the concerns. On the issues of Figure 2A highlighted with the yellow boxes, Pérez-Castiñeira commented more precisely (highlights mine):
“It seems that we made a mistake when we elaborated this figure for the paper. The results shown in this manuscript were part of a Ph.D. thesis that it was going to be submitted in 2010 (although the Ph.D student finally quit, moved to the private industry, and never got her Ph.D.). A pdf with a thesis chapter (generated on April 2nd, 2009) in which you can see the original image (Figure 5 of this chapter) is available at: https://www.dropbox.com/s/7zv7wg22cjzdusk/Complementacion H%2BPPasas en YPC3.pdf?dl=0 You can see that the result was identical to that shown in Figure 2A.“
Here is that Figure 5, Cheshire (aka Actinopolyspora biskrensis) spotted more duplicated panels:



First author and PhD candidate Rocio Drake left the University of Sevilla in 2010 for chemical industry, since 2017 she is a teacher at the Albrecht Dürer German School in Sevilla. Did she quit without graduating because of legitimate concerns about the quality of her data? We’ll probably never know [she ignored my inquiry via LinkedIn, -LS], but please keep this figure in mind because the surprises aren’t over.
Regarding the problems of Figure 4A and S1A, Pérez-Castiñeira stated (highlights mine – A.):
“We were so interested in the growth on glucose (the proof of the functional complementation of yeast soluble PPase by different chimaeric membrane PPases) that we might have made some mistakes with the drops on galactose, where the growth was always identical. We did these experiments so many times with so many different transformants that, at the end, we had tens of plates with tens of spots each one. Galactose plates were particularly “boring” because we never saw any significant difference in growth on this carbon source with any of the constructions tested […] There were three people involved in taking photos and we had images with plates upside up and upside down, therefore, sometimes we had to flip the images and it may have been the case that we made mistakes when we did it. In any case, this was absolutely unintended and does not change the results whatsoever: we have been using YPC3 for nearly 20 years and all of them grow exactly the same […] This was appreciated by many other scientific groups that rapidly asked us to provide YPC3 for their own work. […] None of these group have ever complained about YPC3 phenotype, actually, two of them have published papers that confirm our results with this mutant […]“
It’s remarkable that with “tens of plates” available, three people taking photos were unlucky enough to pick always the same ones, and later no-one spotted the unintended mistakes.
When asked if he intended to inform the journal about those “mistakes” and publish corrected new figures, Dr. Pérez-Castiñeira politely declined the invitation giving these reasons:
“I was not thinking of notifying the journal because, as I told you in my previous message, these mistakes do not mean anything in practical terms. […] Along those years, I guess we took pictures of more than a hundred plates, each of them having an average of 25-30 spots, because we checked the ability of tens of soluble and membrane-bound PPases from different sources to functionally complement the yeast soluble PPases and, of course, we had to make sure that the results were reproducible, therefore, the experiments had to be repeated at least three times. Only a few of these pictures were selected for this particular paper, which means a lot of work for three people and, in my view, increases the probability of mistakes. In any case, I can only guess what might have happened but I cannot be certain about what actually happened. I also want to add that, although we are a small group working on a relatively unknown scientific field, we have our reputation and have published in top journals for more than twenty years without having any problems or complaints. […]“
You see? There’s nothing to be worried about “in practical terms”, just a myriad of of wrong data in three out of six main figures and in one supplementary figure. Wait, it’s not over yet, Cheshire found more in Figure 6:

Our talkative Dr. Pérez-Castiñeira was again keen to clarify:
“These figures were elaborated by me personally and I remember quite well how I made them. Definitely, the images shown in this figure were cut and pasted from the originals in order to elaborate the figures, the overlapping was done “manually” using GIMP and, also, in some cases, brightness and contrast were adjusted.”
Brilliant. Fearless. No qualms in admitting what he did, cut & paste is common practice there, no more questions please. It’s somehow admirable the attitude of Dr. Pérez-Castiñeira in speaking openly without hesitation. Then his comment goes on and on, with lots of detailed information on how they store the yeast strains in fridges, on the originals images stored in several DVDs in his office, “because I think I have hundreds of photographs” he added. Photographs! What kind? Vacation photographs? Did Pérez-Castiñeira mean micrographs maybe? Yeah, I’m nitpicking, and it’s just one single article showing signs of data manipulation after all, isn’t it? Well, not really. Do you remember his PNAS article he quoted to support the data of Drake et. al 2010? Here it is:
Perez-Castineira JR*, Lopez-Marques RL, Villalba JM, Losada M, Serrano A*. Functional complementation of yeast cytosolic pyrophosphatase by bacterial and plant H+-translocating pyrophosphatases PNAS (2002) doi: 10.1073/pnas.242625399


A PNAS paper, likely something not common in 2002 for a small university like Sevilla and especially in plant sciences, which may have meant the tenured professorship for the first author Pérez-Castiñeira. Of note, this article was “communicated” in PNAS by the late Nobel Prize winner and National Academy of Sciences member Arthur Kornberg, meaning the paper was accepted outside of a proper peer-review process.
You might have noticed that the most popular deceiving technique used in the lab of Pérez-Castiñeira’s mentor Aurelio Serrano is anything but sophisticated, and a true trademark: vertical flip. Old school. It’s artisan vintage fraud, the cheaters of today are operating on a much higher level of digital fakery.
Remember I asked you to keep in mind the Figure 5 of Drake’s thesis? There’s a reason why:

The red box highlights the same yeast strip used in Figure 1A of the PNAS article (please check above), where the complementation by IPP1 was in YPC-1 cells, while YPD cells were used in Drake’s thesis. And of course Drake was not author of the PNAS paper, she was still an undergraduate student back then.
It doesn’t seem credible to me that there are that many plates and pictures stored in fridges and on DVDs in Sevilla. Rather, it looks the contrary is true: the same old stuff being recycled over and over again. In case you wonder if our talkative Pérez-Castiñeira commented on these issues too – of course he did. Not immediately this time though, it took nine months after the thread was created on PubPeer. Brace yourself (highlight mine):
“Dear Sir, we have contacted the editors at PNAS and given our explanations to the issues and concerns expressed by you. The editors have supported these explanations and have encouraged me to post this message to Pubpeer. Thanks for your interest in our articles.”
Very short and evasive for his standards. Pérez-Castiñeira & Serrano haven’t spontaneously contacted the editors themselves, most probably. I informed the editors months ago on the case and I assume the editors contacted them, not vice versa. Isn’t it reassuring that PNAS “supported” those explanations, although I informed them that the group has a list of problematic articles? I queried the Editor-in-Chief May Berenbaum and some journal editors for explanations. A few days later I was honoured with the following answer from the editorial ethics manager Ms. Yael Fitzpatrick (highlights mine – A.):
“Dear Aneurus Inconstans, We previously evaluated this paper and found no action was necessary. However, based on your comment we are reevaluating the matter. Thank you for your interest. Kind regards.”
Second chance for PNAS, maybe this time they managed to read my concerned email from start to finish.
Yet another piece of knowledge produced by this incredible lab, with lane duplications in Northern blot analysis:
López-Marqués RL, Pérez-Castiñeira JR, Losada M, Serrano A Differential regulation of soluble and membrane-bound inorganic pyrophosphatases in the photosynthetic bacterium Rhodospirillum rubrum provides insights into pyrophosphate-based stress bioenergetics J. Bacteriol. (2004) doi: 10.1128/JB.186.16.5418-5426.2004


No comments were posted in PubPeer by the group on this case. It was almost disappointing not to read that “those mistakes don’t mean anything in practical terms“, but perhaps it would have been too bold even for Pérez-Castiñeira to give reasons of pure mistake or unintentional oversight this time.
Now a much more recent paper published in 2019, showing an obvious splice, and immediately below you’ll read a funny reply by Aurelio Serrano (highlight mine):
Serrano-Bueno G, Madroñal JM, Manzano-López J, Muñiz M, Pérez-Castiñeira JR, Hernández A, Serrano A* Nuclear proteasomal degradation of Saccharomyces cerevisiae inorganic pyrophosphatase Ipp1p, a nucleocytoplasmic protein whose stability depends on its subcellular localization Biochim Biophys Acta Mol Cell Res (2019) doi: 10.1016/j.bbamcr.2019.02.015


“Thanks for your comment. I will contact the person who did this experimental work, who has not worked in this laboratory since several years. Since it is already Christmas vacations I will not be able to give you an answer until the beginning of next January. Sincerely”
Cheshire replied promptly:
“Thank you for responding. Out of curiosity, do the individuals who perform experiments generally remove the original data from the lab when they leave?”.
Cheshire’s question is more than appropriate, as the paper was published less than two years before Serrano’s reply on PubPeer. Serrano never followed up with his promise to clarify.
Let’s conclude the roundup of scientific irregularities by this plant science group, featuring yeast again:
Pérez-Castiñeira JR, Hernández A, Drake R, Serrano A* A plant proton-pumping inorganic pyrophosphatase functionally complements the vacuolar ATPase transport activity and confers bafilomycin resistance in yeast Biochem J. (2011) doi: 10.1042/BJ20110447
A whole set of problems are present here, ranging from a panel duplication (red boxes), to a repeated portion of a panel (blue boxes), up to repetitive markings (within the magenta box) similar to those created when using a clone tool.
Do you believe that Pérez-Castiñeira refrained from commenting in the face of such tampering? Not at all, he did not lose heart and decided to give us yet another enlightening explanation:
“The hard disk where the originals of these images was accidentally reformatted and they were lost; however, I keep originals of preliminary experiments as well as images of another experiment I did in 2017 with YPC4. These plates confirm the results shown in this paper. […] I attach links to my dropbox account, where I happened to find these old pictures, that belong to an informal talk I gave to my lab mates. As you can see, although YPC4 was published in 2011 we had been working with it much earlier. Please, note that the best rescue of YPC4 phenotype was obtained with chimeric versions of AVP1 that had GFP attached at its N-terminus. These chimeras were generated a bit later and, therefore, are not included in this experiment.” [then a list of dropbox links was provided by Pérez-Castiñeira -A.] “You can use the following figure as a guide to interpret the above attached figures:”

Unfortunately in the lower half of this “guide” figure some yeast strips were duplicated (blue boxes) or duplicated and again vertically flipped (orange boxes).

I hope this story has got you a laugh, and all the credit goes to José Roman Pérez-Castiñeira, without whose comments and “explanations” this report would have had a much duller flavour.

Other Iberian yeast masters
Masterful yeast-art can be also admired in many articles by well-known Spanish plant scientists like Ramon Serrano (Pedro L. Rodriguez’s mentor, now emeritus professor at IBMCP Valencia), Vicente Rubio (group leader at CNB-CSIC in Madrid), Roberto Solano (professor at CNB-CSIC in Madrid), and Salomé Prat (professor at CRAG in Barcelona).
Seven articles by Ramon Serrano have been flagged on PubPeer, but we have never had the privilege of reading any comments on these problems from Prof. Serrano as of today. Here is one such paper in urgent need of commenting:

Vicent I, Navarro A, Mulet JM, Sharma S, Serrano R* Uptake of inorganic phosphate is a limiting factor for Saccharomyces cerevisiae during growth at low temperatures FEMS Yeast Res (2015) doi: 10.1093/femsyr/fov008
Figure 1A, obviously the very first figure of the article. Hard to understand how no-one spotted those “mistakes”, reviewers included.
Another one by Ramon Serrano’s lab published years ago, again it’s quite remarkable that the blatant duplication in Figure 4C went unnoticed:
Mulet JM, Alejandro S, Romero C, Serrano R The trehalose pathway and intracellular glucose phosphates as modulators of potassium transport and general cation homeostasis in yeast Yeast (2004) doi: 10.1002/yea.1126

Then an EMBO Journal paper from 2002, likely something that helped the first author Lynne Yenush (former Serrano’s postdoc and now Deputy Director of IBMCP) to obtain her tenured position:
Yenush L, Mulet JM, Ariño J, Serrano R The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression EMBO J. (2002) doi: 10.1093/emboj/21.5.920

Figure 8A, a set of yeast colonies was duplicated and digitally modified (yellow boxes), as a light dot is present in the background of the picture to the right but not to the left one, the colonies are identical.
Allow me a little digression. Digital modification of yeast colonies is a practice that I’m tempted to believe it has been handed down over time at IBMCP Valencia from Serrano to Rodriguez and then to the young, rising star Julia Santiago, whose supreme masterpiece Santiago et al. 2009 still awaits to be retracted (as promised) by The Plant Journal (as well as other three papers filled with manipulated data in other journals), and whose whole PhD thesis is a collection of forgeries (allegedly committed by someone else).
It seems that Dr. Santiago has so far passed unscathed all the investigations promised by the University of Lausanne, by the ethics committee of the European Research Council (ERC) and the Polytechnic University of Valencia. She has recently published her first paper as group leader in a high profile journal, she is regularly invited to chairing sessions at international meetings, and everything flows nicely without anything untoward happening. Hats off.

Dr. Santiago will chair the Structural Biology session at the International Congress on Plant Molecular Biology, 23-27 October 2022 in Cairns, Queensland, Australia.
Let’s move on to a tribute to Vicente Rubio, a guy we’ve also met before in the Pedro Rodriguez & Julia Santiago story with the problematic Irigoyen et al. 2014 paper, where Rubio was the sole corresponding author. Here some more problematic yeast data authored by his lab:
(left) Cardona-López X, Cuyas L, Marín E, Rajulu C, Irigoyen ML, Gil E, Puga MI, Bligny R, Nussaume L, Geldner N, Paz-Ares J*, Rubio V* ESCRT-III-Associated Protein ALIX Mediates High-Affinity Phosphate Transporter Trafficking to Maintain Phosphate Homeostasis in Arabidopsis Plant Cell (2015) doi: 10.1105/tpc.15.00393
(right) Park J, Lim CJ, Shen M, Park HJ, Cha JY, Iniesto E, Rubio V, Mengiste T, Zhu JK, Bressan RA, Lee SY, Lee BH, Jin JB, Pardo JM, Kim WY, Yun DJ* Epigenetic switch from repressive to permissive chromatin in response to cold stress PNAS (2018) doi: 10.1073/pnas.1721241115


The PNAS paper described right above here is a collaborative work with Dae-Jin Yun‘s lab at Konkuk University in Seoul, South Korea, a guy who has his own PubPeer record with 4 entries. It’s possible that Rubio’s co-authorship was indeed granted due to Figure 1A, given his expertise with yeast-two-hybrid (Y2H). It’s just a negative control, however, I must have read somewhere that one’s research is as good as their controls are.
The following Genes & Development article, with Rubio as first author, could have launched Rubio’s career towards the position of group leader. Unfortunately, clearly manipulated data are recognizable in the Northern blot of Figure 6, the three lanes of the RNA-binding protein4 (RBP4) gene control are the same, beautifully disguised as a continuous blot. One can just imagine how reliable the rest of the paper is:
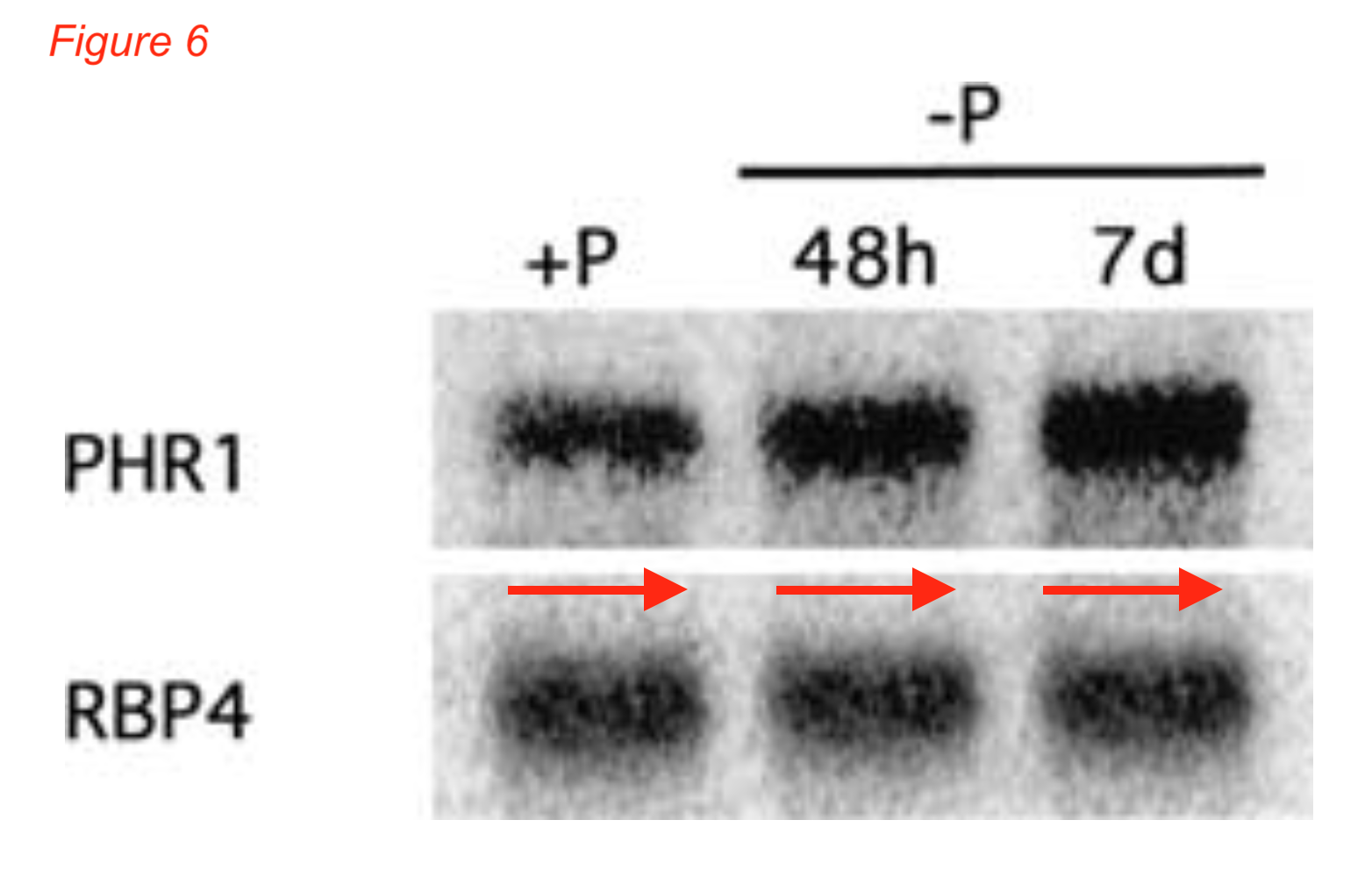
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J* A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae Genes Dev. (2001) doi: 10.1101/gad.204401
Yet another pillar of Spanish pant science, Roberto Solano, who recently published a paper on the phantom anti-COVID properties of the chlorophyll catabolite pheophorbide A in an MDPI (sic!) journal, has a PubPeer record of 6 articles and has his own troubles with yeast data too. In one of these papers you’ll notice the name of Vicente Rubio’s mentor Javier Paz-Ares (4 entries in PubPeer for him), because science is a village:
(left) Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R* (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate Nat Chem Biol (2009) doi: 10.1038/nchembio.161
(right) Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, Pauwels L, Witters E, Puga MI, Paz-Ares J, Goossens A, Reymond P, De Jaeger G, Solano R* The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses Plant Cell (2011) doi: 10.1105/tpc.110.080788
It seems that the reuse of negative controls often happens in Solano’s lab. Isn’t peculiar that the same kind of mistake occurs over the years in the same place?
Now a collaboration between Solano and Carlos R. Figueroa‘s lab from University of Talca, Chile, three colonies have been reused to describe different interactions (and I have doubts on a fourth pair of colonies, but resolution is too low). The way in which these images have been cropped and rotated makes me sceptical that these errors were accidental:
Garrido-Bigotes A, Valenzuela-Riffo F, Torrejón M, Solano R, Morales-Quintana L, Figueroa CR* A new functional JAZ degron sequence in strawberry JAZ1 revealed by structural and interaction studies on the COI1-JA-Ile/COR-JAZs complexes Sci Rep (2020) doi: 10.1038/s41598-020-68213-w


And finally, after a long list of male PIs, let’s partially rebalance the gender equality index with Salomé Prat, another bigwig of Spanish plant science, with 5 entries in PubPeer where yeast problems are almost always involved. Just one example here, in the high profile journal Current Biology, in its Figure 1A (!):
Nieto C, López-Salmerón V, Davière JM, Prat S ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex Curr Biol (2015) doi: 10.1016/j.cub.2014.10.070

As you can read from the caption of the figure, these are rather significant errors. I do seriously doubt Dr. Prat will check the data and eventually correct those significant mistakes published in a high profile journal, as she has previously refused to correct a mistake, that she acknowledged to be one, in a Nature (!) paper of hers. Here is the article, below you can read Dr. Prat point of view on the matter (hyperlink and highlights mine):
de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S* A molecular framework for light and gibberellin control of cell elongation Nature (2008) doi: 10.1038/nature06520

“Products obtained by semiquantitative PCR amplification of a chromatin immunoprecipitation experiment with PIF4 are shown in this Figure. DNA gels are less subject to running deformation than protein gels and thereby uninended panel duplications are difficult to notice by naked eye. Image shows 8 negative amplifications out of 12 tested promoters and so, it is evident that it was not manipulated on purpose. It had been just as simple as omitting this particular gene in the final Figure. We went back to the lab notebooks and a copy of the original gel is included below. [low resolution raw data were here provided – A.] It can be observed that in fact we duplicated by error this image. This mistake however affects a negative result and does not invalid the experiment and the interpretation of data. We regret this mistake but believe it is not strictly compulsory asking the Journal for publication of a correction note, including the ammended Figure.“
In a country, where dishonest scientists are viciously defended by their elite peers and where critics of research fraud are forced to publicly apologise, it is intriguing to see both Dr. Prat and Dr. Pérez-Castiñeira refusing to correct their own acknowledged mistakes.
I thank all my donors for supporting my journalism. You can be one of them!
Make a one-time donation:
I thank all my donors for supporting my journalism. You can be one of them!
Make a monthly donation:
Choose an amount
Or enter a custom amount
Your contribution is appreciated.
Your contribution is appreciated.
DonateDonate monthly



The “zombie” paper Santiago_et_al_2009 got already 15 citations this year so far, and 41 citations in 2021. Considering that the PubPeer thread was opened in June 2021, we can assume the paper got agout 20 citations in the second half of 2021.
In other words, this highly fraudulent article got cited 35 times since the issues were highlighted on PubPeer and notified to the journal. AND the corresponding author P.L. Rodriguez asked to retract the article, but nothing has happened.
The Plant Journal editorial board informed me it will take a little longer to “conclude the investigation”.
Here are the issues, judge for yourselves whether it’s acceptable that such investigation could take that long, and especially after the corresponding author asked to RETRACT the paper:
Figure 1:
Zoom ins of Figure 1:
Figure 3B, a lovely rectangular patch:
Figure 4D:
LikeLike
Cardona-Loper et al. 2015 was corrected on March 28, 2022:
https://academic.oup.com/plcell/advance-article/doi/10.1093/plcell/koac103/6555007
LikeLike
Irigoyen et al. 2014 was also corrected on March 28, 2022:
https://academic.oup.com/plcell/advance-article/doi/10.1093/plcell/koac102/6555005
LikeLike
Dupeux et al. EMBO J. 2011 (by Rodriguez & Santiago) was corrected on April 4, 2022:
https://www.embopress.org/doi/full/10.15252/embj.2022110799
LikeLike
Yet another problem in Figure 2A of Perez-Castineira et al. 2011, another strip was reused and flipped vertically to desribe different constructs (yellow boxes):
https://pubpeer.com/publications/DF63F425206425A593483DF132DACB#6
LikeLike
Expression of Concern (EoC) for Drake et al. 2010 Biochem J.:
https://portlandpress.com/biochemj/article/479/14/1517/231583/Expression-of-Concern-N-terminal-chimaeras-with
LikeLike
“The Editorial Office has been made aware of potential concerns surrounding the scientific validity of this paper, hence has issued an expression of concern to notify readers whilst the Editorial Office investigates.”
LikeLike
Retraction for Perez-Castineira et al. 2002 PNAS:
https://www.pnas.org/doi/full/10.1073/pnas.2213841119
Figure 1B was also completely made up:
https://pubpeer.com/publications/E508D13C9E57C1592418405681BD9D#8
LikeLike