Smut Clyde, with the help of Tiger BB8, uncovers another paper mill scam in China. The count is currently at ~100 publications.
Unlike in previous cases, where the “authors”, or rather the customers were hospital doctors (or even mathematicians) without any labs or research funding but with a need to churn out some research papers, the situation here is different. These paper mill customers are active Chinese biomedical researchers, in high academic ranks of professors and deans, and recipients of some very serious governmental research grants. Only that the cancer and neurology research they publish in allegedly respectable journals, is utterly fake, the figures of flow cytometry, immunohistochemistry, western blots and even mixed-up organ photos being provided by a paper mill and recycled between various customers. When caught, the “authors” blame a “third party” and play victims.

The fungal entities have acquired forepaws and might be sneezing sphinxes
By Smut Clyde
This is Wednesday so it must be time for another Papermill Exposé, with accompanying spreadsheet.
Now no-one wants ForBetterScience to become an all-Papermill channel. And we cannot really expect to shame or inspire scriveners in the academic-ghostwriter industry to seek out more constructive applications for their talents, so the point of exposing them is not immediately obvious. Since papermills are a natural response to ill-thought-out government incentives and pressures (intended to increase a country’s research productivity, as measured by publication count), the situation will only change when governments change those policies… until then, we can only provide material for journalists and science advocates in those countries. Spotting and characterising the mills is also a chance to practice one’s skills in the traffic analysis and EOB branches of SIGINT, or so I hear from a friend.
A novel feature here is a couple of Mea Tulpas* in which authors describe how things went went tits-up after they outsourced parts of the research to unnamed contractors. They are as surprised and appalled as the original PubPeer contributor by the use of manipulated images, and have promised to retract the papers in question. This is a reminder that the papermills covered previously at FBS, which fabricate papers in their entirety and even handle correspondence with the journals, are only the extreme end of a spectrum.
Certainly there is room in the fake-science ecology for services who complete half-written manuscripts and flesh out the listed authors’ partial results with their own, making the manuscripts more publishable with figures that are well-composed even if unrelated to the nominal topics of research. It seems, though, that the clients take a gamble when they send their work off for completion: assuming that these contractors have the facilities to conduct the crucial experiments, why would they bother? And what is there to stop them copying your own images to on-sell to future customers?
Authors
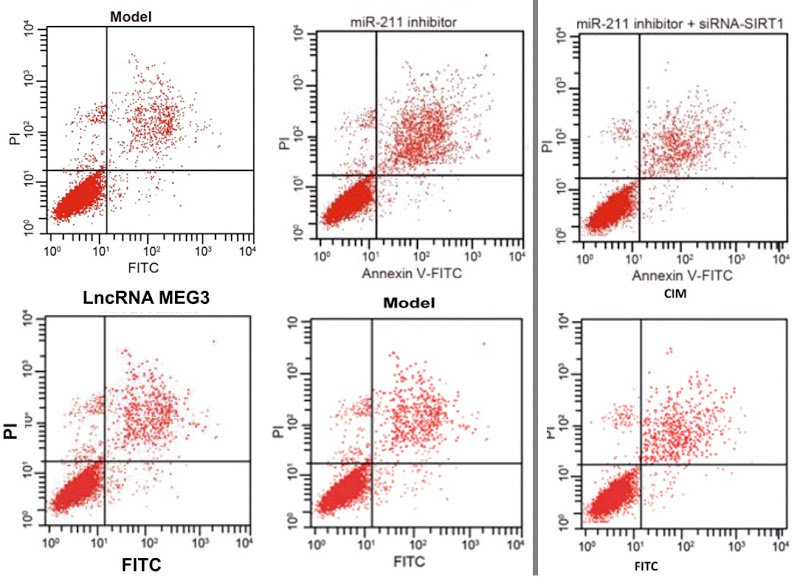
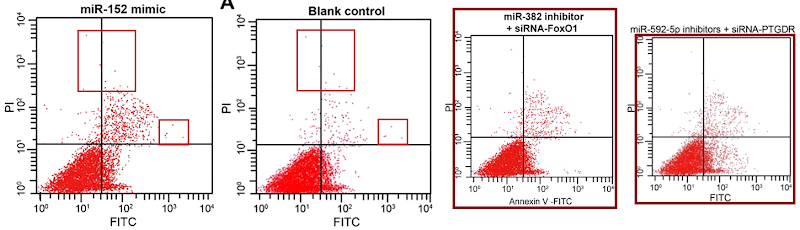
My sympathy goes out to Pan et al (2018) [21] and I hope they get their money back, but it is a pity that they do not name the third party responsible for this chicanery, so that others could benefit from the sad experience. The PubPeer discussion in this case related to Flow-Cytometry apoptosis results, in which the levels of two fluorescent protein markers are measured in individual (dyed) cells as they stream past a laser, and used to locate the cells as points on a scatterplot. Or six scatterplots to illustrate how different non-coding-RNA treatments affect cell viability.
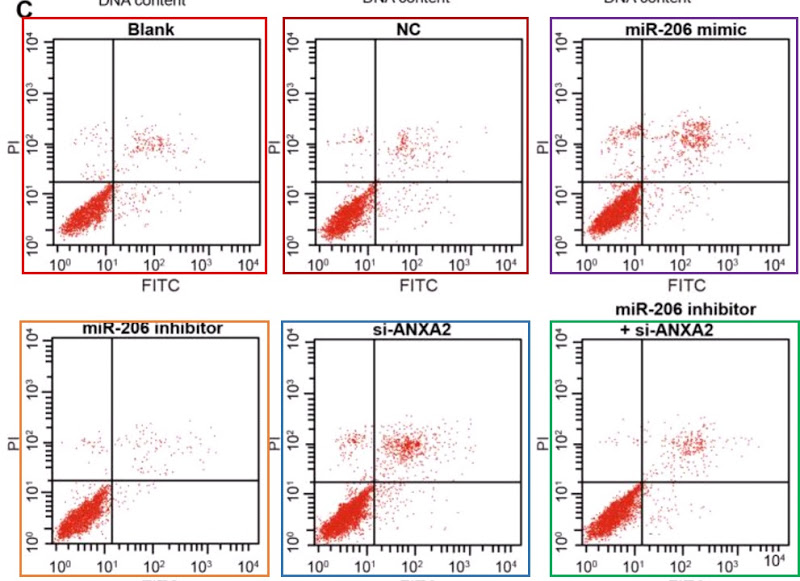
As well as selling these apoptosis results to Pan’s group, the unscrupulous millers fobbed them off on Zhang et al (2017) [30] (twice, as Figures 10A and 10C)…
…and on Li et al (2018) [14]. Corresponding panels are not identical, for each version has been customised with an additional stippling of dots, but the amount of duplication in the upper-left and lower-right quadrants is dispositive.
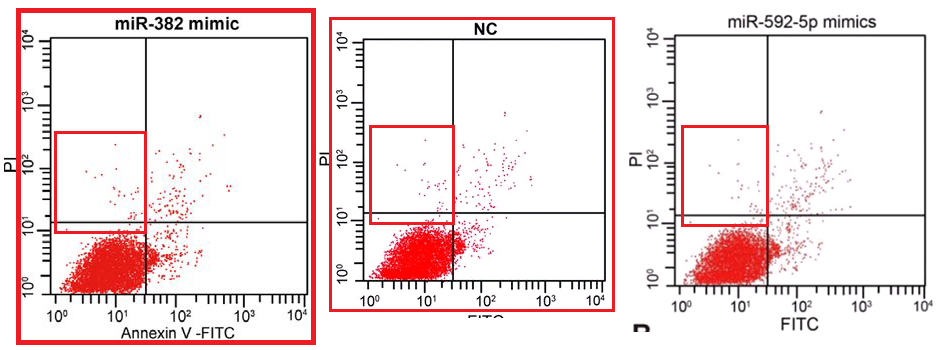
Scatterplots abound in this oeuvre, with the lower-left quadrant occupied by a metaphorical firework mortar, or the proverbial smoking gun, or a champagne bottle; or an exploding puffball spraying a cloud of spores into the upper-right quadrant. It may be that these metaphors dominated the designers’ minds, causing them to arrange the dots in radiating streaks (almost as if the coordinates are not sampled randomly from a distribution but otherwise uncorrelated). The firework displays are frequently more similar than one might expect, within and between papers.
Five papers are illustrated with versions of a second cluster of champagne-bottle plots. Five more papers draw on a third cluster, in which the fungal entities have acquired forepaws and might be sneezing sphinxes. Sphinges (because 3rd declension)? More than one sphinx.
Other panels within this third cluster will always be “Spiny Norman” to me.

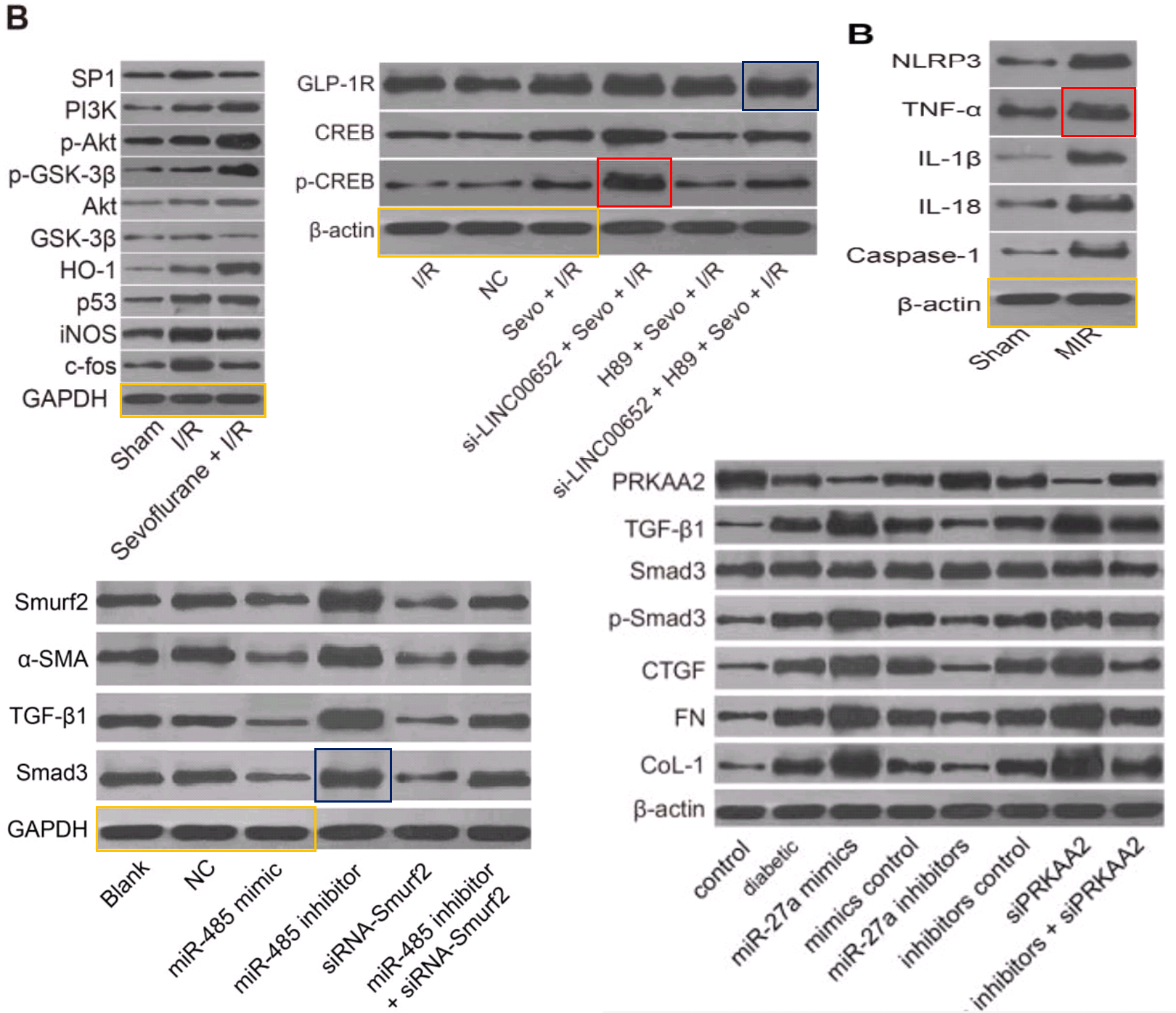
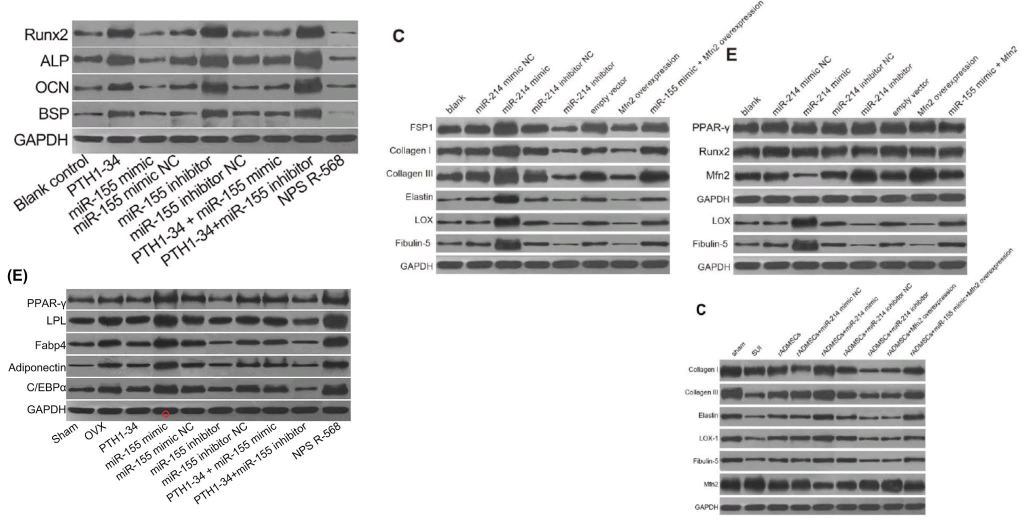
The corpus under present scrutiny comprises some 30 80 90 100 papers. They are linked by a number of recurring features which together create a family resemblance. In addition to the Apoptosis flaw-cytometry plots, [14] and [21] exemplify a concourse of Western Blots where the protein blobs all emerge from a restricted gamut, like letters of an unfamiliar alphabet strung together in different sequences… though with loading-control bands that repeat from paper to paper. These dominate the corpus and we’ll come back to them.
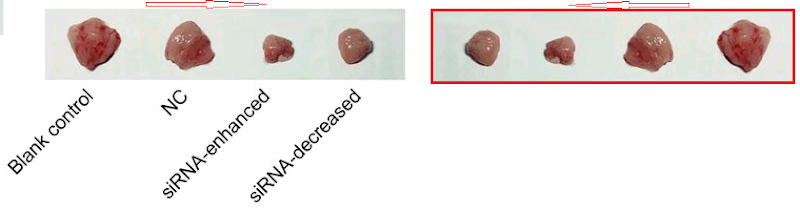
At the other extreme, [30] opens a door into a small gallery of tumors with only three six sightings at the time of writing. The grid at left, Fig 14B of Lu et al (2018) [27], are purportedly human B16 melanoma xenografts. Some are duplicates, including pairs of smaller tumors that aren’t marked because I ran out of colored pencils. The emphasis is on the reappearance of the same visceral postage stamps at right – flipped horizontally or rotated through 180° – in Fig 12B of [30], and now human osteosarcoma xenografts (cell-line unspecified).
Another application of four of these tumors identified them as esophagal cancer (Fig 7C of Wen et al (2017) [15]).
Added in proof: Figs 2B from Wang et al (2017) [29] and 6D from Wu et al (2018) [31]


Or perhaps they were liver carcinoma xenografts, as in Fig 11A from He et al (2018) [5].
All this is to say that the papers in this corpus vary more, with less of a cookie-cutter production-line quality than the papermill products previously explored at FBS and by Dr Elisabeth Bik at Science Integrity Digest. There are more idiosyncratic, bespoke image manipulations. This is consistent with the working hypothesis that they are collaborations between the studio and the individual customers. Of course this makes it harder to spot examples with certainty, because there is no single sufficient diagnostic feature! Some entries in the spreadsheet are bound to it by only one or two common threads, and it may be that any given peripheral member does not belong; the falsified images could have been faked independently, or by a rival contractor.
The period covered is basically a two-year window. Which is not to say that the contractors ceased to operate after 2018: they may have removed themselves from the radar by evolving to a different repertoire of methods and images. Five delayed cases from 2019-2020 all appeared in European Review for Medical & Pharmacological Sciences, and I am inclined to suspect that another papermill which targets that journal acquired material from the main studio.
A range of journals are involved, some more hospitable to bogosity than others. Cellular Physiology & Biochemistry is disproportionately represented with 39 56 entries (runners-up are the Journal of Cellular Biochemistry and Journal of Cellular Physiology, both from Wiley, with 11 each). Until recently CPB was published by Karger, who ran down its value with their focus on publishing fiction, to the point that a learned society based in Düsseldorf could buy it out. One purpose of this post is to increase those researchers’ workload as they slowly go through the journal’s back-catalog dealing with the more egregious dreck.
Some were for repeat customers, with the two papers from Bao-Long Pan’s group where we began. Four entries in the spreadsheet from Shu-Bo Zhang et al at the Affiliated Hospital of North China University of Science and Technology. A group at the Key Laboratory for Biotechnology on Medicinal Plants of Jiangsu Province, School of Life Science of Jiangsu Normal University (Xuzhou, Jiangsu) – evidently helmed by Dong-Mei Wu, Jun Lu and Yuan-Lin Zheng – are especially loyal, with 30+ entries. I’ll come back to them.
As well as [14], Zhi-Feng Li commissioned another paper which was submitted only eight days later to the same Bioscience Reports journal, but with a different burner email address (for the same corresponding author), suggesting that at least one was sent directly by the scribes at the third-party contractor. Both papers are illustrated with either squamous-cell carcinoma cells (A431 cell-line) or carcinoma cells (B16 line) infiltrating through a Matrigel filter after diverse treatments… the images being the same, mildly photoshopped.
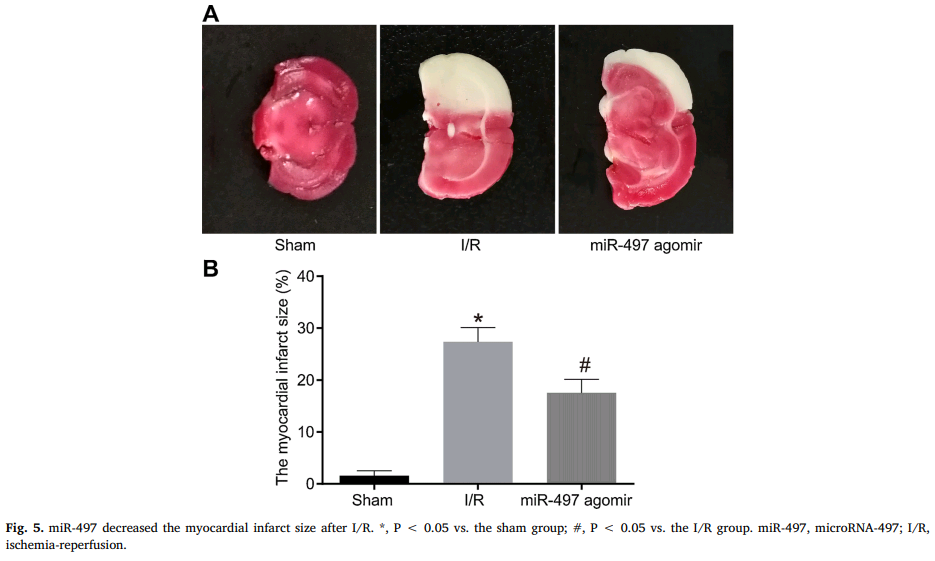
The priority now is the contents of this compendium of papermill productivity! Several enlivening aspects commend it to our attention. The use of slices of stroke-damaged rat brains to illustrate claims about treating cardiac ischemia – not once in Zhang et al (2018) [24], but again in Qin et al (2018) [11] – hints at a deep-rooted Head / Heart confusion, and sows the seeds of doubt about the standards of peer review at CPB and at Biomedicine & Pharmacotherapy, the other journal responsible.
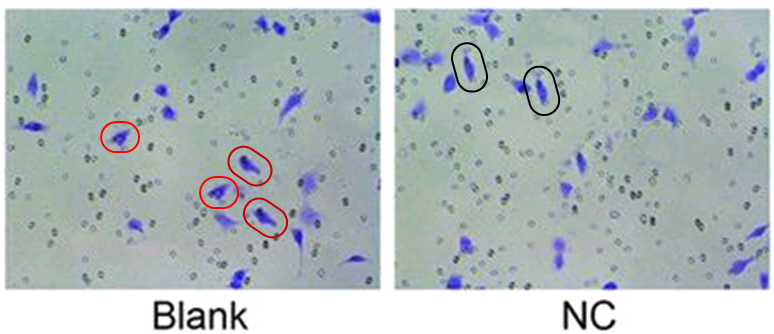
The Head / Heart confusion continues in Wu et al (2018) [26], where a paragraph describes the effects of different gene-modulating micro-RNA treatments on muscle-fibre density in the striatum. This is all very well until you reflect that the corpus striatum is a region of the frontal lobe where musculature is not a good thing.
“The effects of miR-200a and cAMP on the striatum structure in the PD rats were explored by observations under an electron microscope. In the normal rats, the neuromuscular junctions in the striatum were arranged neatly, the structures of the presynaptic and posterior membranes were intact, and the structures of the vesicles and mitochondria were complete. Compared with the normal group, the other six groups had different degrees of changes, such as dense muscle fibers, disordered arrangement, increased presynaptic membrane vesicles and mitochondria, and postsynaptic membrane folds. There were no differences in the above indicators among the blank, NC and miR-200a inhibitor + Forskolin groups (all p > 0.05). Compared with the blank group, the miR-200a inhibitor group had a slightly decreased muscle fiber density, slightly intact arrangement, unremarkably increased presynaptic vesicles and mitochondria and decreased postsynaptic membrane folds, whereas the miR-200a mimic and Forskolin groups exhibited more muscle fibers, more disordered arrangement, more presynaptic membrane vesicles and mitochondria and more wrinkled postsynaptic membranes (Fig. 5). Altogether, the inhibition of miR-200a or the inactivation of cAMP could improve the striatum structure in PD rats.”
Joining the conversation in the PubPeer thread, the authors blamed the error on the pressures of publishing in a language in which they are not fluent. Unable to match the elegance of descriptions elsewhere, they copy-pasted them, without noticing that the wording was inappropriate for brains.
What brings [26] under the papermill’s aegis is Fig 5, a polyptych of TEMs, illustrating neuronal responses to those chemical challenges.
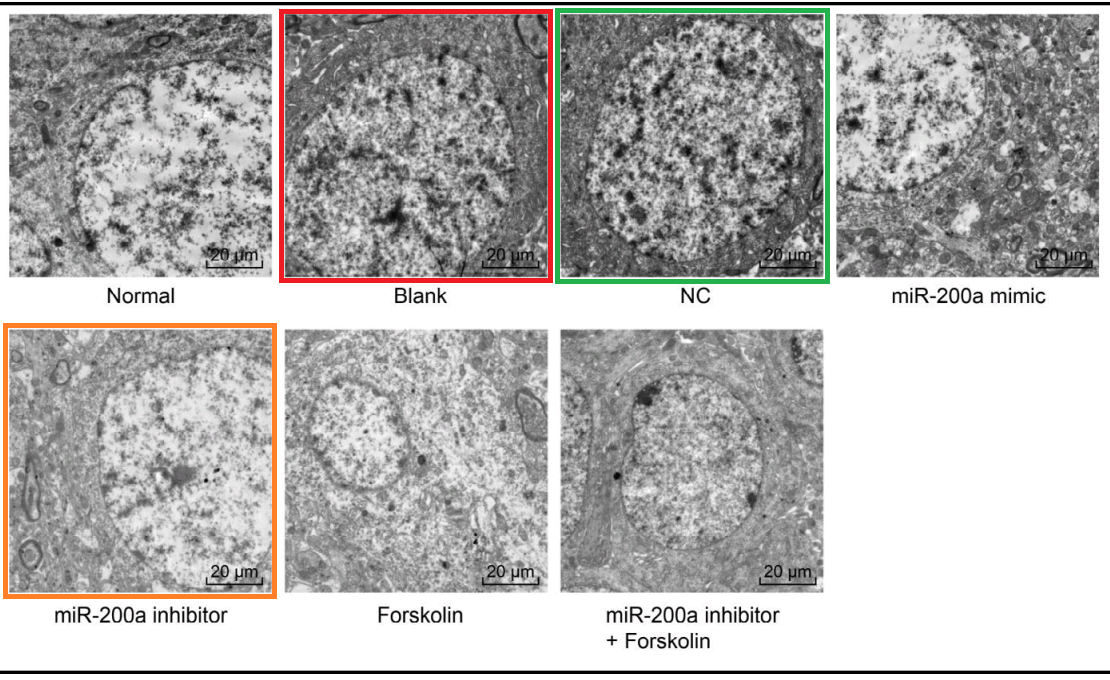
It resonates with five similar polyptychs in which the electron-microscopy targets are usually the somata of hippocampal neurons – specifically pyramidal neurons in Wen et al (2018) [6] – but the cell-lines of breast carcinoma in Zhang et al (2016)[18] and Gu et al (2018) [23]. Teams of authors are non-overlapping. How does this happen?
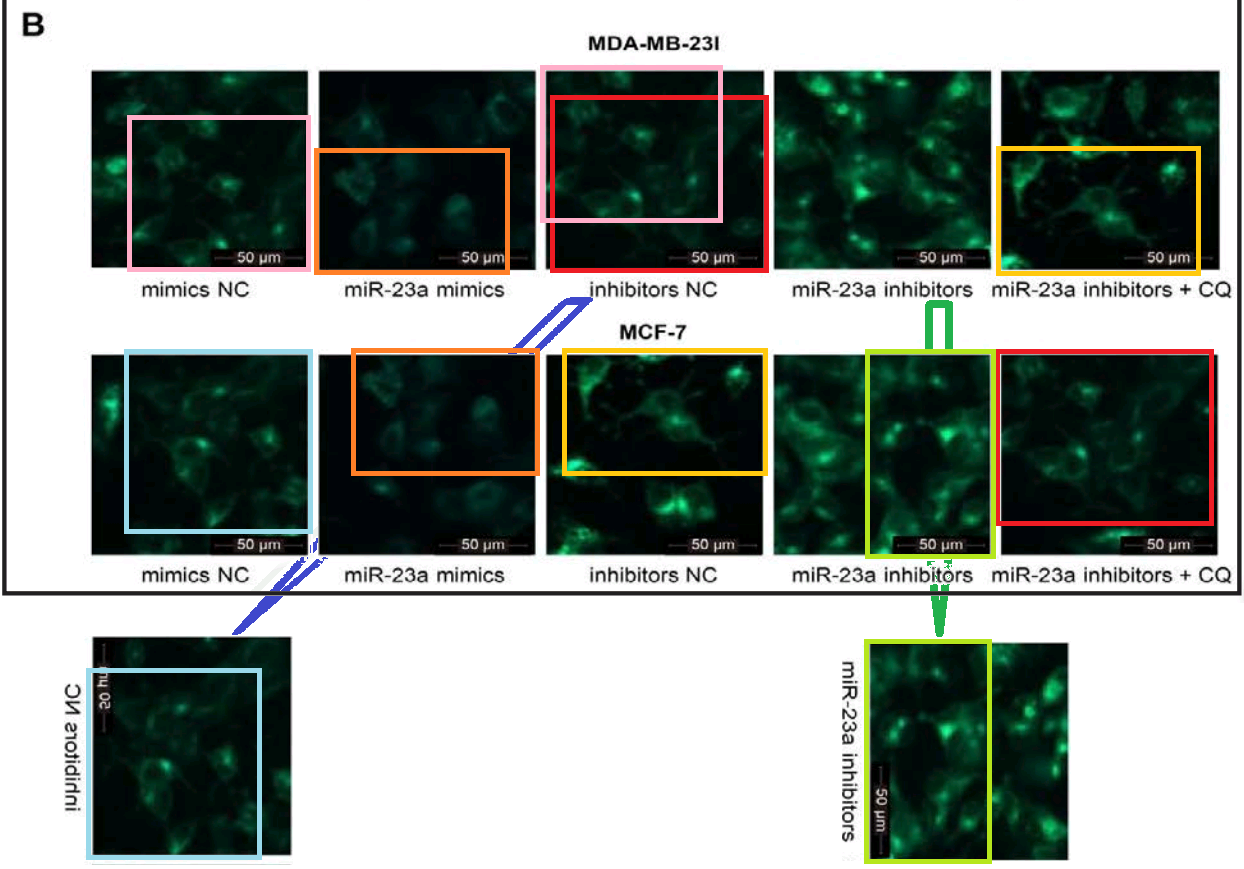
Fig 3D from [23]: “GAS5 inhibits autophagy in breast cancer cells … (D) Transmission electron microscopy was used to detect autophagy in MDA-MB-23l and MCF-7 cells after overexpression or down-regulation of GAS5”.

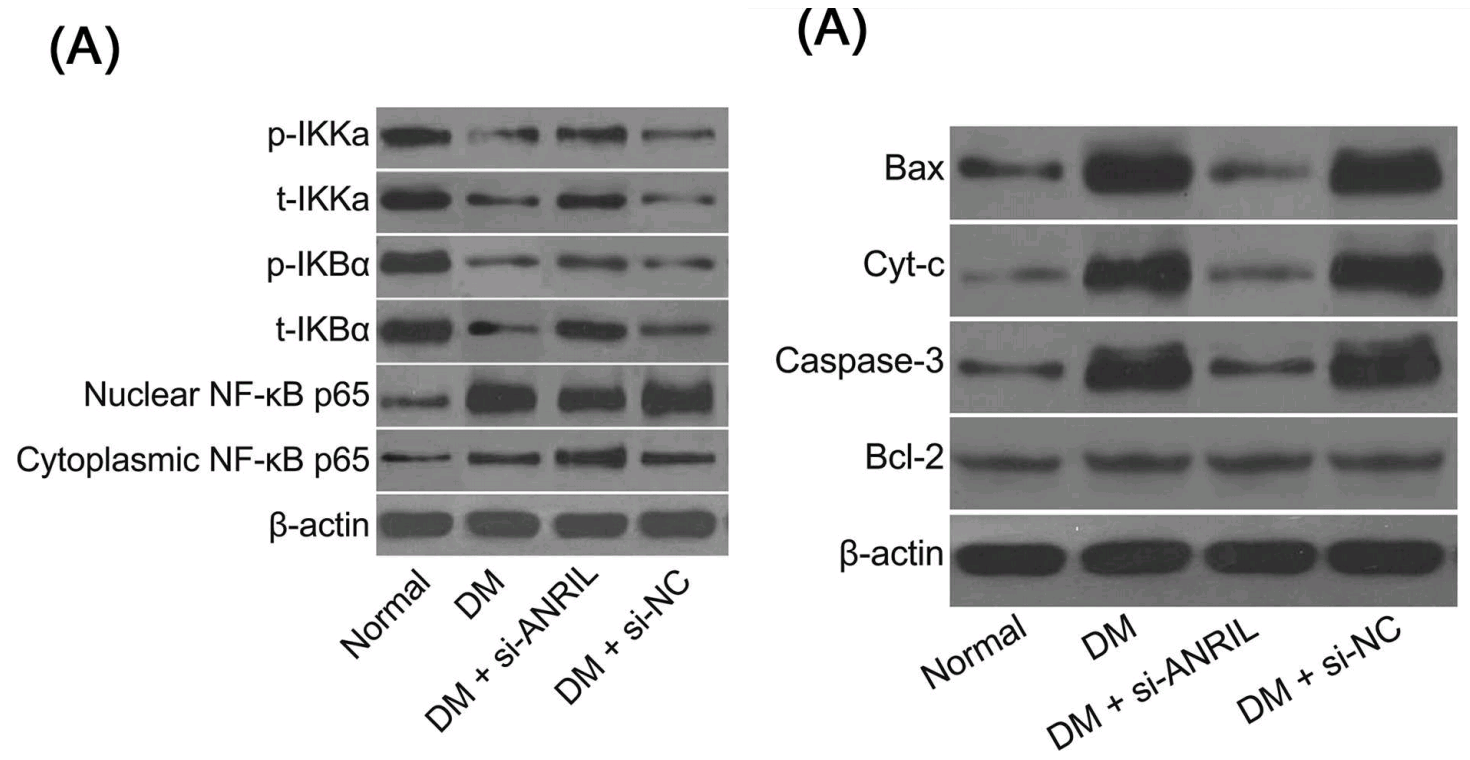
Fig 9 from [6]: “Ultrastructure of pyramidal neurons in the hippocampal CA1 region observed under the transmission electron microscope among the normal group, DM, DM + si-ANRIL, DM + si-NC groups”.
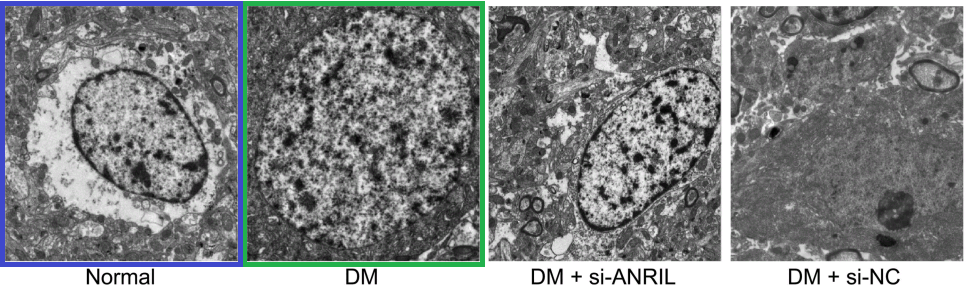
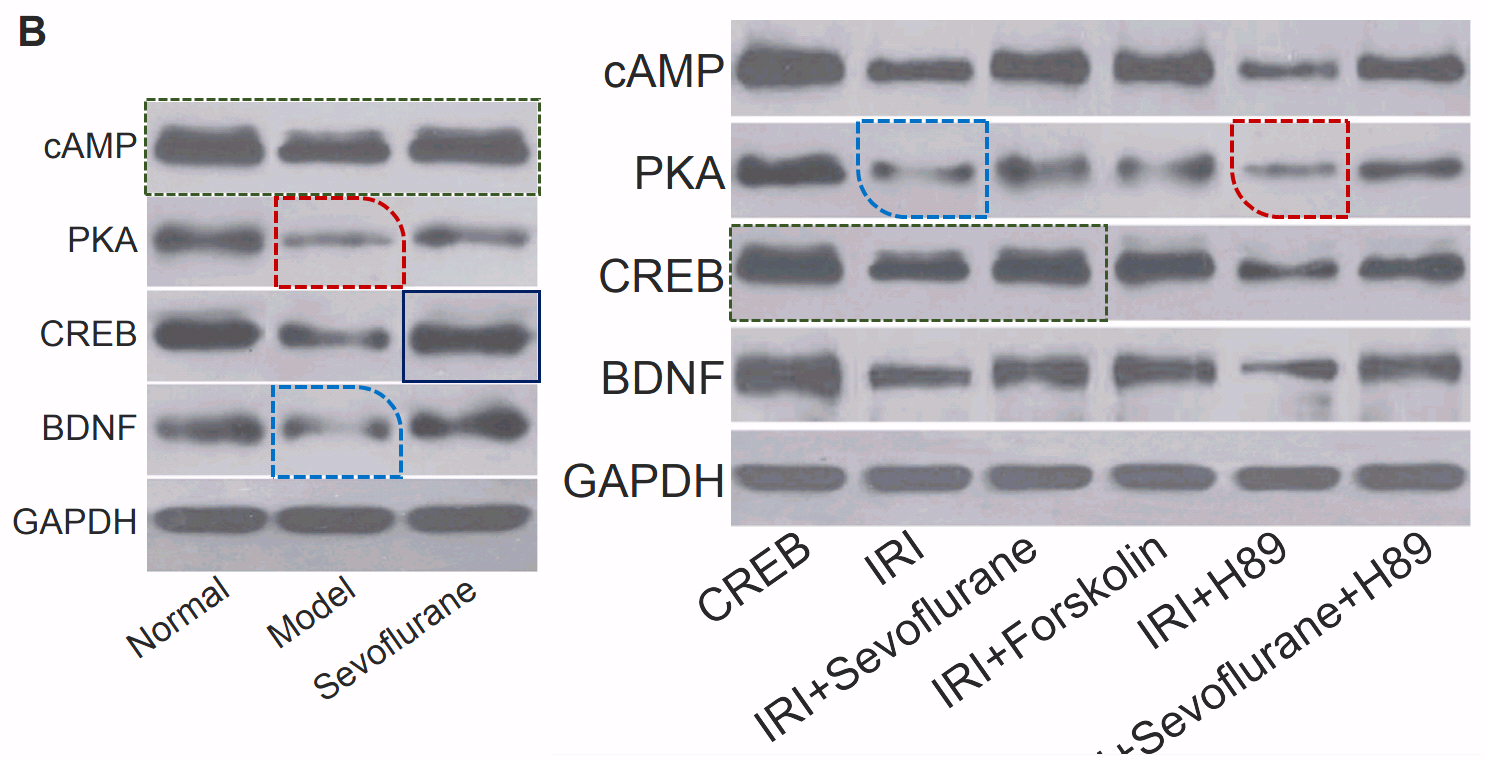
Fig 1 from Liu et al (2018) [13]: “Morphology of hippocampal neurons in the normal, the model and the sevoflurane groups”.

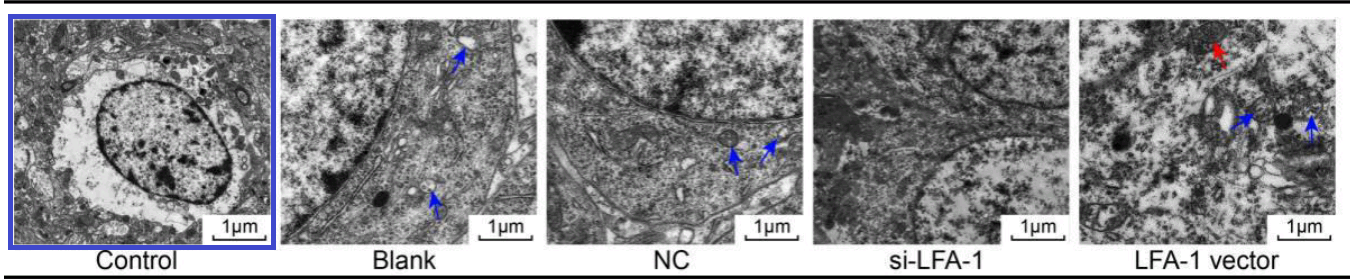
Fig. 2 from Yu et al (2018) [25]. “LFA-1 gene silencing improves ultrastructure of hippocampal neurons in CA1 area.”
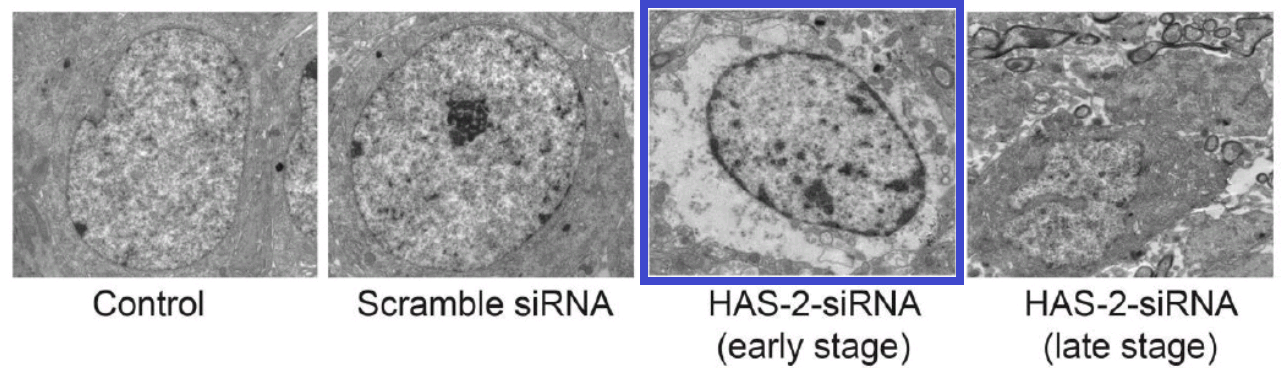
This thread began with 2016, when Figure 2 of [18] showed “Effects of HAS-2 gene silencing on the ultrastructure of MCF-7 cells after the transfection of all groups”.
[23] was originally flagged by PubPeer contributor ‘Indigofera Tanganyikensis’ (aka Morty), who admired the economy with which five microphotographs had been reframed and rotated in Fig 5B to illustrate ten different conditions.
[6] and [13] tapped into that vein of alien-alphabet WBs and loading controls, still to be described in detail.
The Acknowledgements in [13] thank the research subjects for their selfless commitment to Science. Laboratory rats so seldom receive the credit they deserve for their sacrifices.
Acknowledgments
We would also like to thank all participants enrolled in the present study.
It would be helpful if Acknowledgements sections also included a note of which papermill had provided the manuscript, or which consultants the authors had paid to adorn their own manuscript with additional figures and results. But I digress.
Overlapping brain sections provide a slightly farther-reaching connection among papers. Fig. 3 of [25] hints at this, though the overlap is purely between panels. “LFA-1 gene silencing ameliorates pathological changes in hippocampal CA1 area of brain tissues, observed by HE staining”.

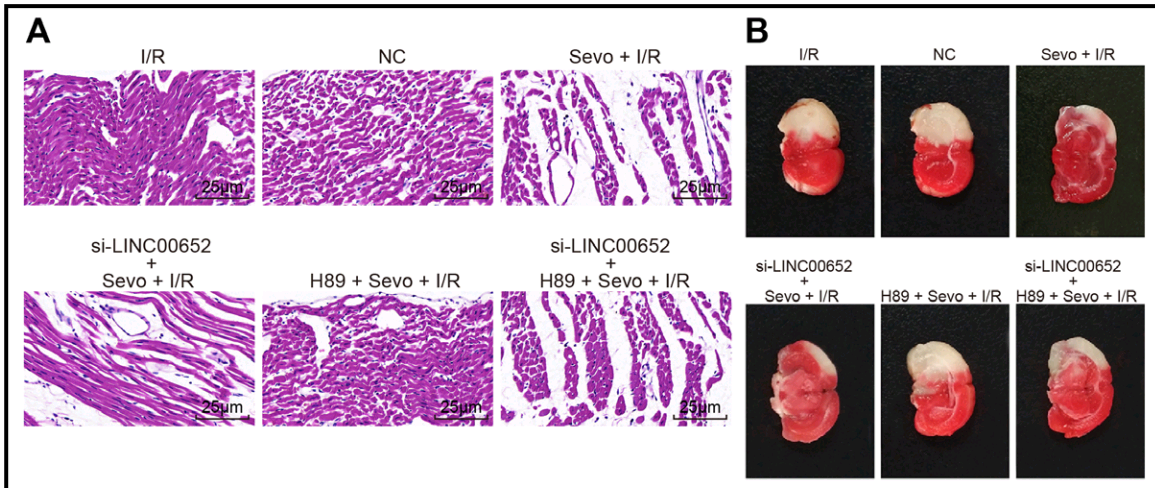
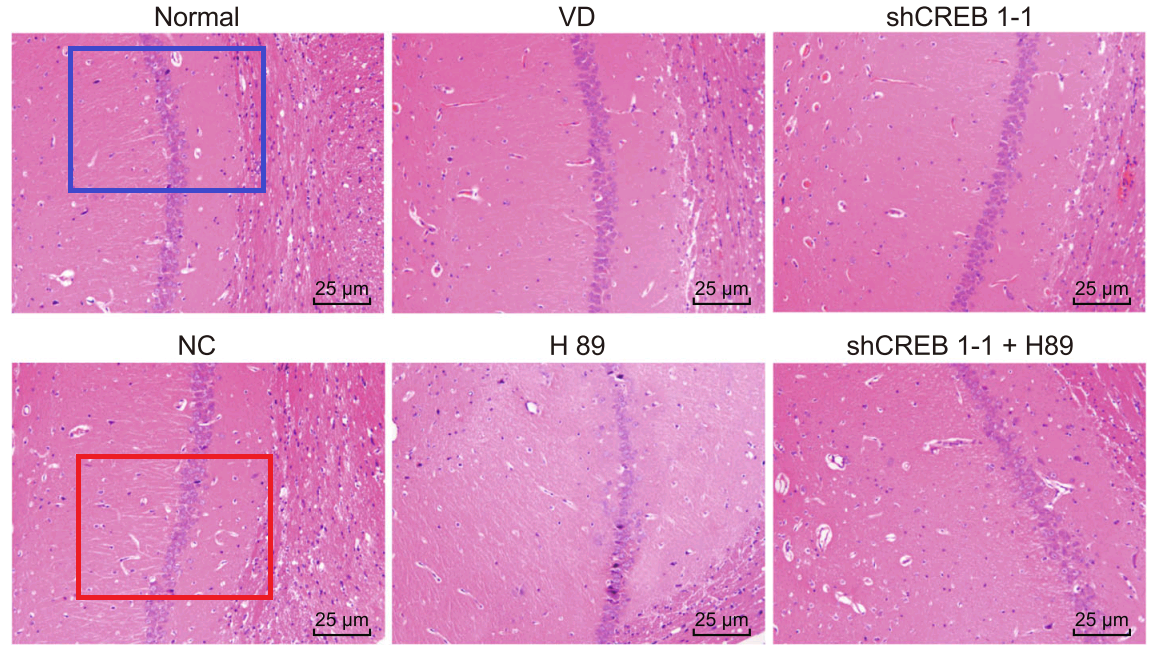
Han et al (2018) [28] leads further afield, with Fig. 3: “Pathological changes of hippocampal neurons of mice injected with shCREB1–1 and H89 are most serious”. One link leads to Fig 3 of Wu et al (2018) [9]: “Pathological changes of hippocampal neurons of mice injected with shCREB1–1 and H89 are most serious”.

In another direction we find Fig. 3 of Piao et al (2018) [22]. “The morphological changes in rat hippocampus cells from each group by HE staining”.


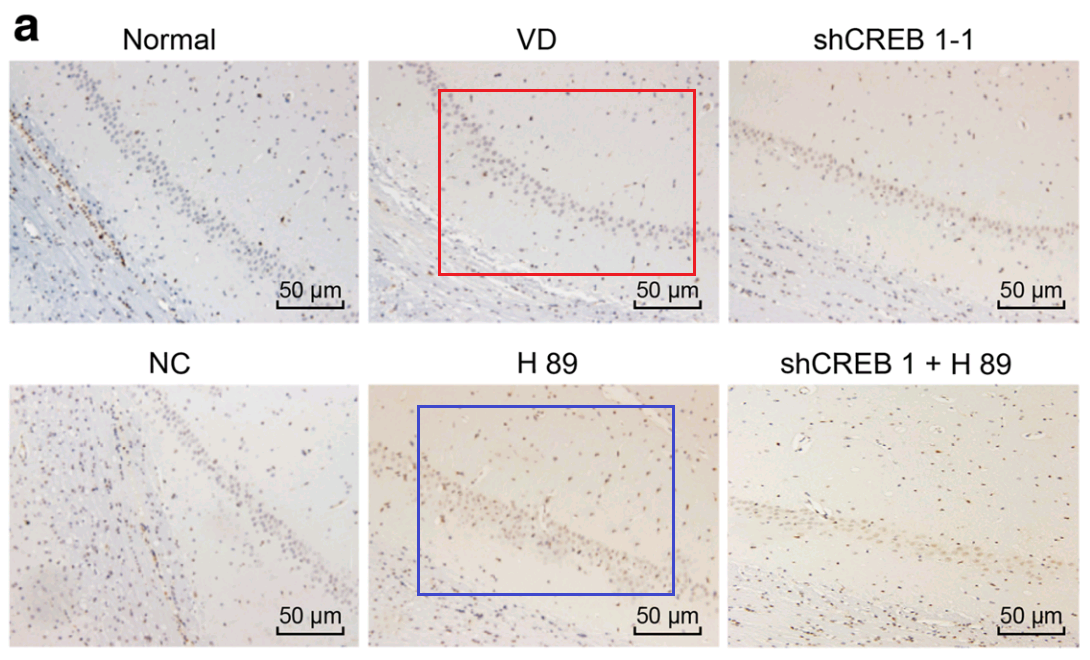
Or one could focus on Fig 6A of [28], “Increased neuronal apoptosis in hippocampal CA1 area of mice injected with shCREB1–1 or/and H89, Notes: A, the TUNEL staining results of CA1 in hippocampus of mice in each group “. It connects with Fig. 7A from Jin et al (2017) [12]: “Apoptosis of hippocampal nerve cells among each group detected by TUNEL assay. Note: A, images of hippocampal nerve cells apoptosis in rats detected by TUNEL assay among each group”.
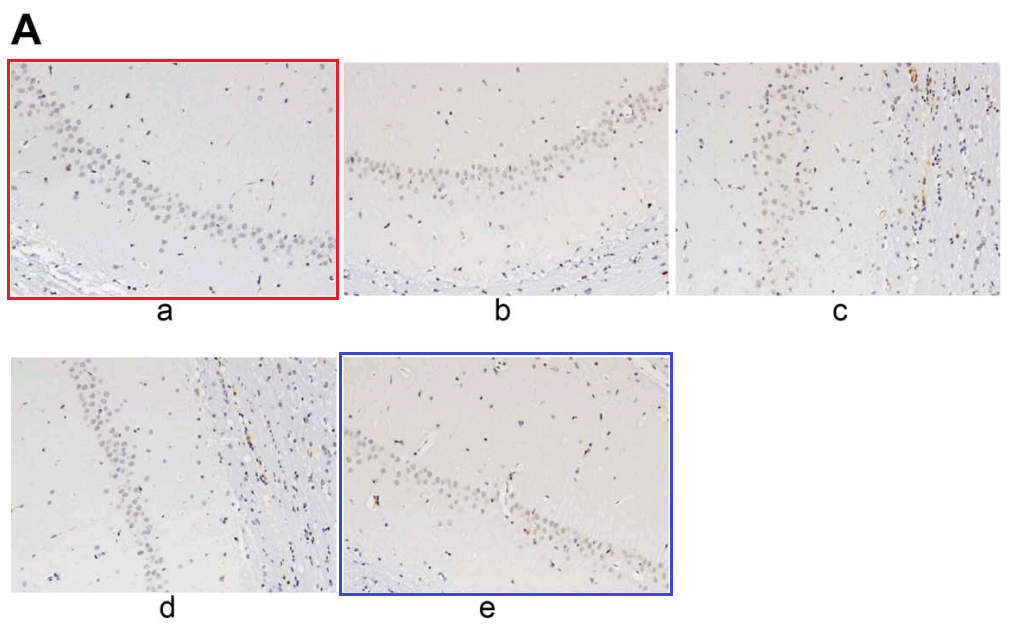
From [12], another panel of 7A overlaps with Fig 2A from Sun et al (2018) [20]. “Neuronal apoptosis in the normal and HIBD groups by TUNEL staining. Notes: A, cell apoptosis in both groups under microscopy”.
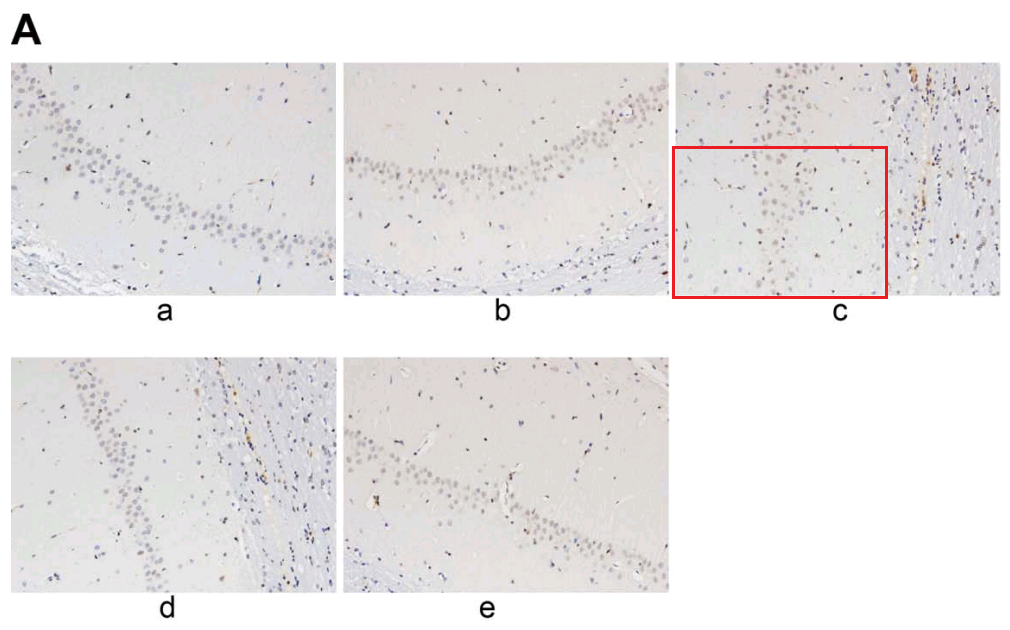
Not to forget the link between Fig 4A of [22]: “Cell apoptosis in rats from each group by TUNEL staining (200 ×) (n = 6). Notes: A, Results of TUNEL staining in each group”, and Fig 4(a) from Wen et al (2018) [7]: “TUNEL staining indicates that the model group had increased cell apoptosis rate in hippocampal CA1 region. Note: (a) cell apoptosis of the two groups using TUNEL staining”.
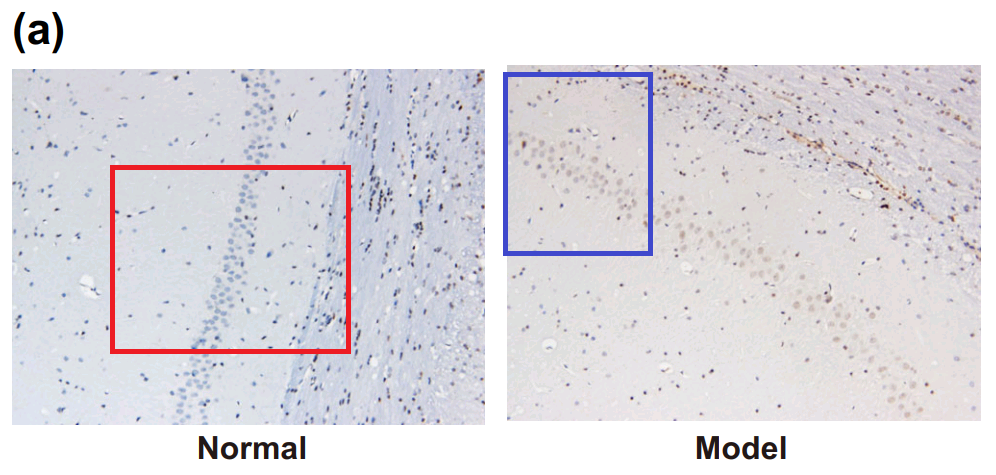

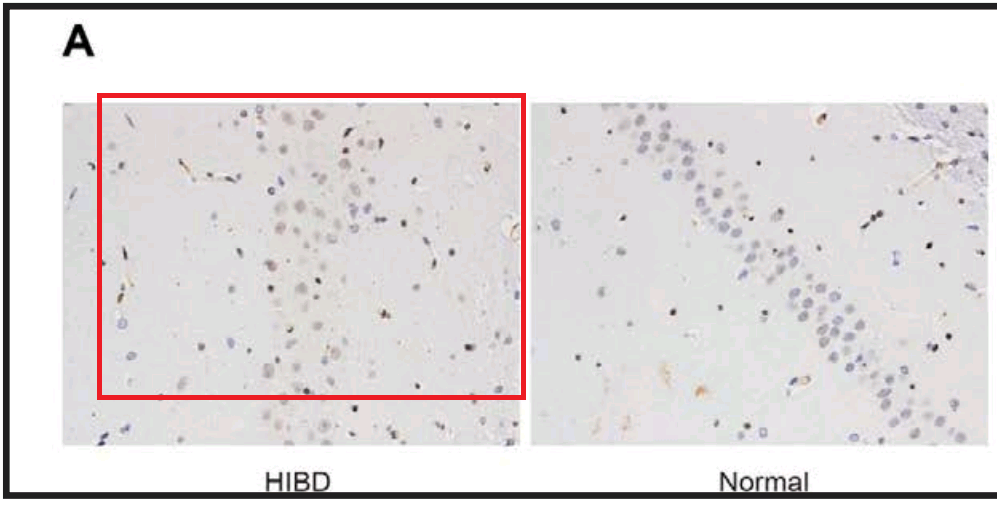
By now readers will have grasped the general picture, so I’ll finish this section with Fig 3A from Han et al (2018) [17]: “The combined action of miR‐140‐5p and DEX protects neurons of hippocampal CA 1 region in the left hemisphere observed by HE staining and Nissl staining. Notes: A, HE staining of pyramid neurons in the hippocampal CA 1 region of the left hemisphere”. One panel can also be found as Fig. 1 of [20], “HE staining of hippocampal tissues in the normal and HIBD groups”.
Another panel recurs in Fig 2(a) from [7], “Pathological changes of hippocampal CA1 region were found in the model group. Note: (a) hippocampal CA1 region in the two groups using HE staining”.
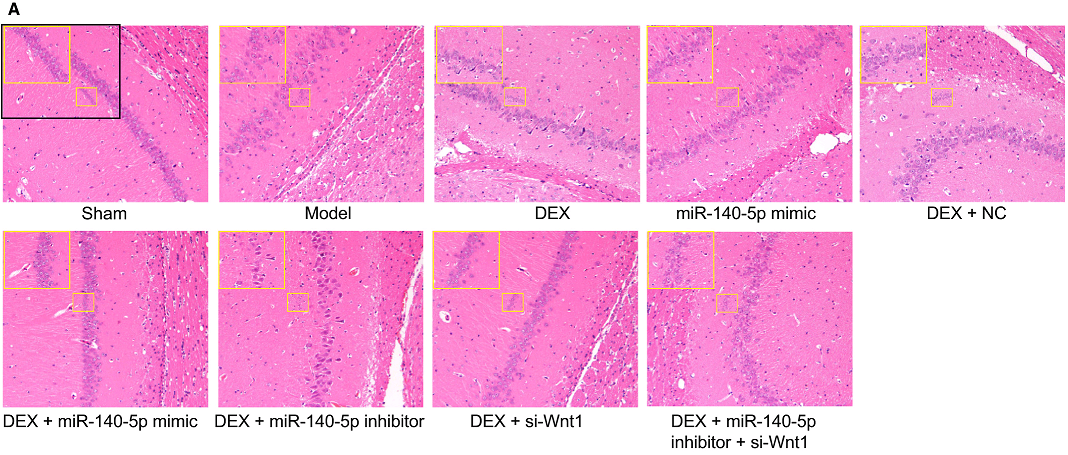

But I lied about finishing yet, because [7] also has Fig 2(b) (“hippocampal CA1 region in the two groups using Nissl’s staining”), while [17] has Fig 1A, and the Normal and Model panels and the NC and Inhibitor panels respectively need only a vertical flip and a 180° rotation to match.
Another possible entry into the corpus is Han et al (2018) [3], originally flagged by Dr Bik for using one image to illustrate two conditions.

This could happen to anyone and is not a grievous fault, but the paper was subsequently retracted because concerns had been brought to the publisher AND the authors lost confidence in their statistical analysis.
The retraction has been agreed following an investigation based allegations raised by a third party, and after the authors stated that due to statistical errors the conclusions of the articles were no longer valid.
This explanation from Wiley Periodicals, in which the accumulation of reasons did not cohere into clarity, did not mention a third potential concern… an illustrative Figure 2C had already appeared, twice, as Fig. 2B of Wu et al (2017) [19], and Fig 3(a,b) of Lv et al (2017).
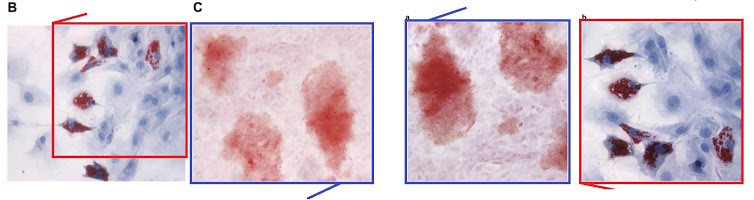
Those earlier papers came from unrelated authors but were under editorial review concurrently, so the duplication was not a simple, straightforward case of plagiarism. Experimental & Molecular Medicine, the journal hosting [k], comes to us from the Nature stable. [19] is from CPB again and showcases Fig 9, a peggy-squared quilt of Photoshop manipulation.
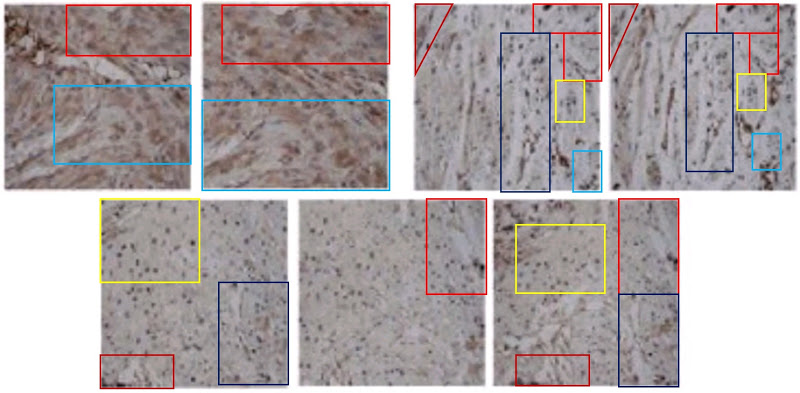
Even without Fig 9, the place of [3] and [19] in the present Wall of Papermill Shame was assured when they drew on that limited alphabet and the same loading controls to compose their Western Blots.
As noted, these ransom notes are rearrangements of a small recurring repertoire of loading controls. Everything is a projective test to me and what I see in one of them is strips of bacon (ten sightings, not all flagged in PubPeer threads).
The “Beds” loading control (eight sightings)…

The “veil”, in which films of ectoplasm are poised above a pair of unsuspecting blobs (ten sightings).


The “canoes” are less popular, with four sightings (including an appearance as a substantive band).
The same is true for the “adze” (five appearances)

But the most versatile loading control in this oeuvre is distinguished by the ‘eye’ on one blob and the little bubble beneath another. Some authors liked those features so much they used the blobs twice in a single band. Figs 4B from [29], 3A from Shen et al (2017) [10], and 4A from Wen et al (2018) [4].


The first and antepenultimate author of [4] were good sports and joined the forensic game at PubPeer, blowing up images from the PDF to show that if you magnify enough, and mis-align the rectangles slightly, you can find differences between image-compression, low-resolution artifacts in different versions – proving that any likeness is merely coincidence. It would have been easier to show this by reproducing the original scan of the electrophoresis gels at full resolution, but I agree that this would have been less fun.


One connecting thread remains. Sometimes it is useful to know the effect of some miR treatment on the phases of the cell cycle: what proportion of the time do cells in a population spend dividing, or focused on tissue-related cell-metabolism tasks undistracted by thoughts of making more cells, or unpacking their chromosomes and duplicating the DNA, or packing up their chromosomes in the aftermath of division? Flow Cytometry enters the picture again, acquiring that information by measuring the level of a single DNA-related protein, to accumulate a protein-expression distribution. The distribution is fitted as a combination of phase-specific templates (one template for the distribution of a population of cells in S phase, one in M phase, and so on). The fitted outcomes inevitably resemble churches with large steeples, and I amuse myself by imagining how Feininger would have painted them as part of his ‘Gelmeroda’ series.
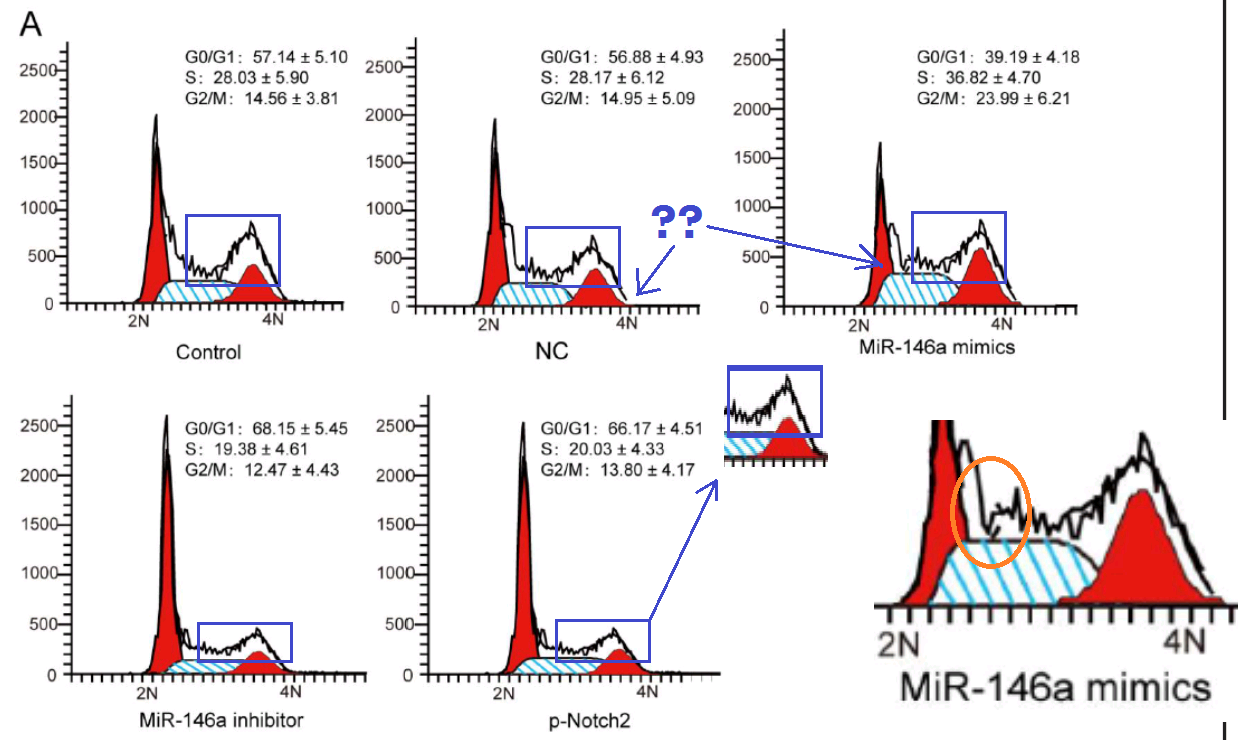
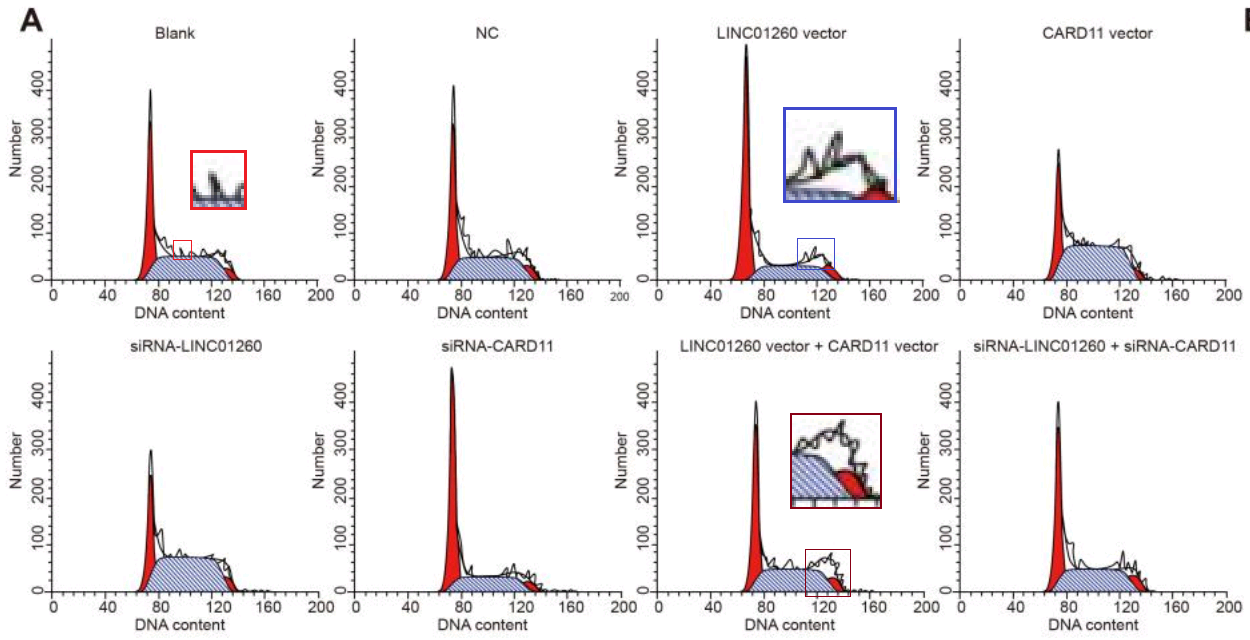
 The key point is that the raw data are a histogram of cell counts graphed against PI levels. Histograms should not overhang like pot bellies or lean like the skyline of Pisa or candles melting near a fire. The peaks should not jut out sideways like the teeth of a circular saw, nor drag their way up the side of the steeple under the impression that they are the Hunchback of Notre Dame.
The key point is that the raw data are a histogram of cell counts graphed against PI levels. Histograms should not overhang like pot bellies or lean like the skyline of Pisa or candles melting near a fire. The peaks should not jut out sideways like the teeth of a circular saw, nor drag their way up the side of the steeple under the impression that they are the Hunchback of Notre Dame.

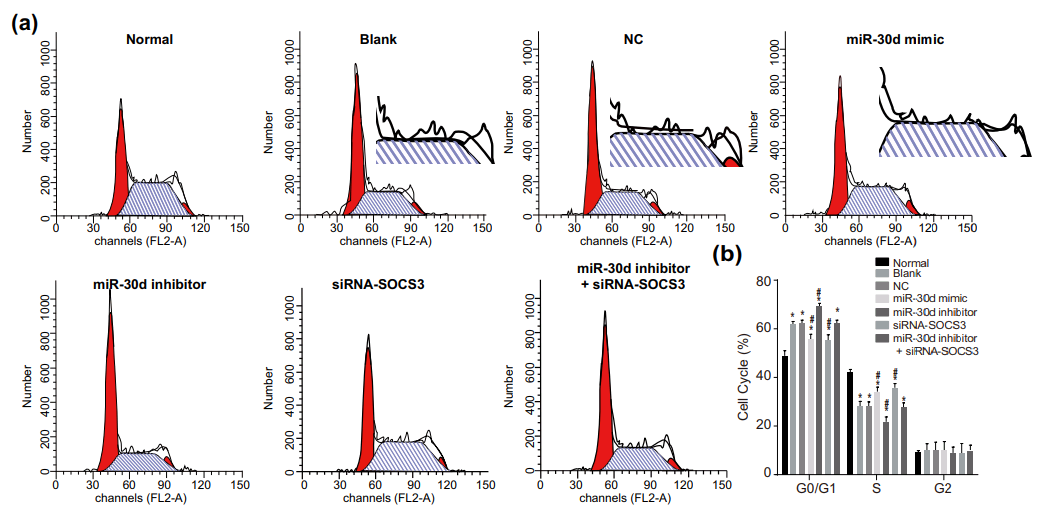
It would also be a surprise if they replicated across multiple experiments or between different researchers’ studies.


In particular, steeple diagrams in four papers follow the same architectural design: J.-Q. Zhang et al (2018) [1], S.-B. Zhang (2018), Qian et al (2018), Wang et al (2018). [1] was the first in this cluster, available on-line before the other three were submitted, and in a response to PubPeer last author Chen-Hong Peng was as shocked as we are by the perfidy and unscrupulosity of some people in academic publishing.
All these interconnecting strands of imagery are at the same time visual clues that indicate when papers were enhanced by this hypothesised papermill / collaborator. This inspires the question that no-one was asking: When a papermill seems to be the likeliest explanation for similarities within a group of papers, how best to find more examples? Sadly, reverse-image searches are not as helpful as one might expect.
Verbal similarities exist. Experience shows that the PubMed indexing entry for a given (e.g.) ‘Bubble-&-Eye’ or Cytometry Steeple paper is likely to include several equally-confabulated products within its list of ‘Similar Papers’. That is, the verbal congruence is obvious to whatever algorithm PubMed uses to measure similarity (unless it is just that most papers are bogus in this Madlibs genre of Non-coding-RNA + cell-messaging-pathway + medical-condition). It would be helpful if the contractors signed their work with a tic or distinctive, diagnostic error, comparable to the ‘Sch-tic’ of a maths mill or the fabled Begger’s Funnel Plot of a meta-analysis production line of yore.
https://platform.twitter.com/widgets.js
However, there is a tension between encouraging readers to join the crowd-sourced Glitch-hunt by showing how easy it can be, versus letting the contractors themselves know how to improve their operations and better cover their tracks. We can be confident, anyway, that if a hundred examples of their faked contributions have come to light, then hundreds more are still waiting in the same journals (and others). If someone wants to use the contractor candidates in the spreadsheet to train a neural network to spot them, go wild! Please don’t train a neural network to write junk papers.
Several entries in the spreadsheet were previously flagged at PubPeer by (inter alia) Xylocampa Areola, who has a nose for papermills; Parashorea Tomentella; Actinopolyspora Biskrensis (aka Cheshire); Dendrodoa Grossularia (I should acknowledge them as well as Dr Bik and Indigofera Tanganyikensis). Which is to say, the papers’ fictitious quality could be seen without needing to compare across papers for recycled material or to train one’s own neural network to spot a distinctive style. Just not seen by reviewers and editors.
These are not low-profile papers that vanished into the digital oubliette of a write-only journal. People are reading them approvingly (or at least finding the titles through G**gle), and incorporating the made-up results as foundations of their own research and summaries of the Current State of Knowledge. Per G**gle Scholar, citations per paper range from 1 to 66 around a mean of 12.2. We can only hope that although the ostensible authors paid an unacknowledged outsider to finish each paper with forgeries, their own research contributions were impeccable.
They are not marginal, funding-starved researchers either!
Quite different from the previous papermill clusters we have discovered, 62 of the 112 papers i.e. 55% of them acknowledged grant support. Twenty-three of the grants were from the prestigious National Natural Science Foundation of China (NSFC), a total of 14.57 millions yuan of RMB. NSFC grants are awarded to highly rated proposals from high level researchers of the entire nation and are considered the top-notch grant funding, similar to an R01 grant from the NIH in the US. The substantial number of NSFC grants involved in purchasing products from this papermill is unprecedent. Previous papermill products rarely listed any grant support, not to say NSFC grants. Eight of the 23 NSFC grants were awarded to the same institution, Jiangsu Normal University in the city of Xuzhou, Jiangsu province. Frankly to say, JSNU is not the front runner in the nation’s top universities in teaching and research, neither the stellar ones even within Jiangsu province. Therefore, a group of 8 NSFC grants likely wasted on purchasing publishing shaky papers is a big deal. These 8 grants may be 10% of the total NSFC grants that this university is able obtain. Six of these 8 grants were from the same group of authors that are affiliated with the Key Laboratory for Biotechnology on Medicinal Plants of Jiangsu Province within the University’s School of Life Science, where the lead author, Professor Yuan-Lin Zheng was the dean until 2008 and his frequent co-author Professor Jun Lu remains the associate dean. Zheng is also the vice president of the university.
There are 15 NSFC grants from other universities involved in this batch of papermill papers. Several of them are from the top tier universities, such as Zhejiang University, Sun Yat-Sen University, and Shanghai Jiao Tong University. Two NSFC grants received by Shi-Qing Feng at Tianjin Medical University had the largest amount of funding, a total of 5.3 million Yuan of RMB. One of this two grants was for promoting international collaboration between China and other countries. Another grant specifically supporting joint research between China and Russia also was reportedly spent on one of the papermill papers published by authors from Tianjin Med University. The funding agency, however, was not NSFC but the Ministry of Science and Technology of China.
In addition to the 23 NSFC grants, there were at least 51 provincial or regional grants used to support the research reported in these papers. For most of them, there is no publicly available information to get an idea of their funding strength; however, there were two grants by the city of Shanghai standing out to be quite substantial, both to the same group of authors led by Dr. Yong-Bing Wang, a surgeon at the Shanghai Pudong New Area People’s Hospital, located in one of the most actively developing industrial area where the thriving hi-tech business make daily progress. These 2 grants to this hospital and one of them was specifically made to Dr. Wang contributed a total of at least 1.5 million yuan of RMB. The 1+ million yuan grant (2015-B-18) was for establishing a key clinical specialty in the hospital, but now we know, at least a small amount of the money aiming to building or improving the hospital’s surgery department ended up in the pocket of the papermill owners.
I outsourced those last paragraphs to a third-party contractor Tiger BB8, so if the numbers turn out to be unfounded in reality I accept no part of the blame.
As already notes, this Key Laboratory on Medicinal Plants was a regular customer. Their output went from six papers in 2016 to 30 in 2017 and 37 in 2018, then down to 22 in 2019 and only eight in 2020 so far (per ResearchGate), with the papermill products concentrated in that 2017-2018 peak of productivity. A triumvirate of names recurs within the authorship lists for these papers, and it is conceivable that one of them needed h-index enhancement to help secure a grant or a promotion or tenure. I am happy to believe that all the minor authors were students or minions who weren’t in a position to question the scale of the outsourcing. I am also happy to believe that frequent main author Y.-L. Zheng accepted co-authorships without thinking to inquire deeply into details of the research, for he was already University Vice-President and had nothing to gain from these papers (which have brought him nothing but aggrievation).
One of these papers [31] was submitted to EBioMedicine, and resided until recently on SSRN, a preprint platform administered by the publishers Elsevier. I write in the past tense because Elsevier have a generous interpretation of publishing ethics; if a preprint becomes an embarrassment to the journal or the authors, it can be wished into the cornfield by withdrawing the submission, leaving its dedicated permanent DOI as a forlorn fingerpost pointing into the void. This happened here:
Just saying, it might be a good idea to perform the relevant studies before strengthening them.
Anyway, the papermill was in operation well before 2017 and is probably still serving customers now, though with the Western Blot random notes written in different alien alphabets, and with a different repertoire of flow cytometry plots (and perhaps with more sophisticated parameters for customising them). So the coterminous nature of the Key Laboratory peak activity and the papermill’s visibility is probably a coincidence.
All these apoptosis raspberries have given me a Shamen earworm from 1989.
* ‘Mea Tulpa‘ is a term from Tibetan Buddhism, meaning “I didn’t do it, but I do have a psychic-projection doppelganger who might have been the culprit”.
Spreadsheet List as Google Docs file
Sources
- “MicroRNA-300 promotes apoptosis and inhibits proliferation, migration, invasion and epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway by targeting CUL4B in pancreatic cancer cells”. Jia-Qiang Zhang, Shi Chen, Jiang-Ning Gu, Yi Zhu, Qian Zhan, Dong-Feng Cheng, Hao Chen, Xia-Xing Deng, Bai-Yong Shen, Cheng-Hong Peng (2018). Journal of Cellular Biochemistry doi: 10.1002/jcb.26270 [PubPeer].
- “MicroRNA-184 inhibits proliferation and promotes apoptosis of human colon cancer SW480 and HCT116 cells by downregulating C-MYC and BCL-2“. Yong-Bing Wang, Xiao-Hui Zhao, Gang Li, Jun-Hua Zheng, Wei Qiu (2018). Journal of Cellular Biochemistry doi: 10.1002/jcb.26330 [PubPeer].
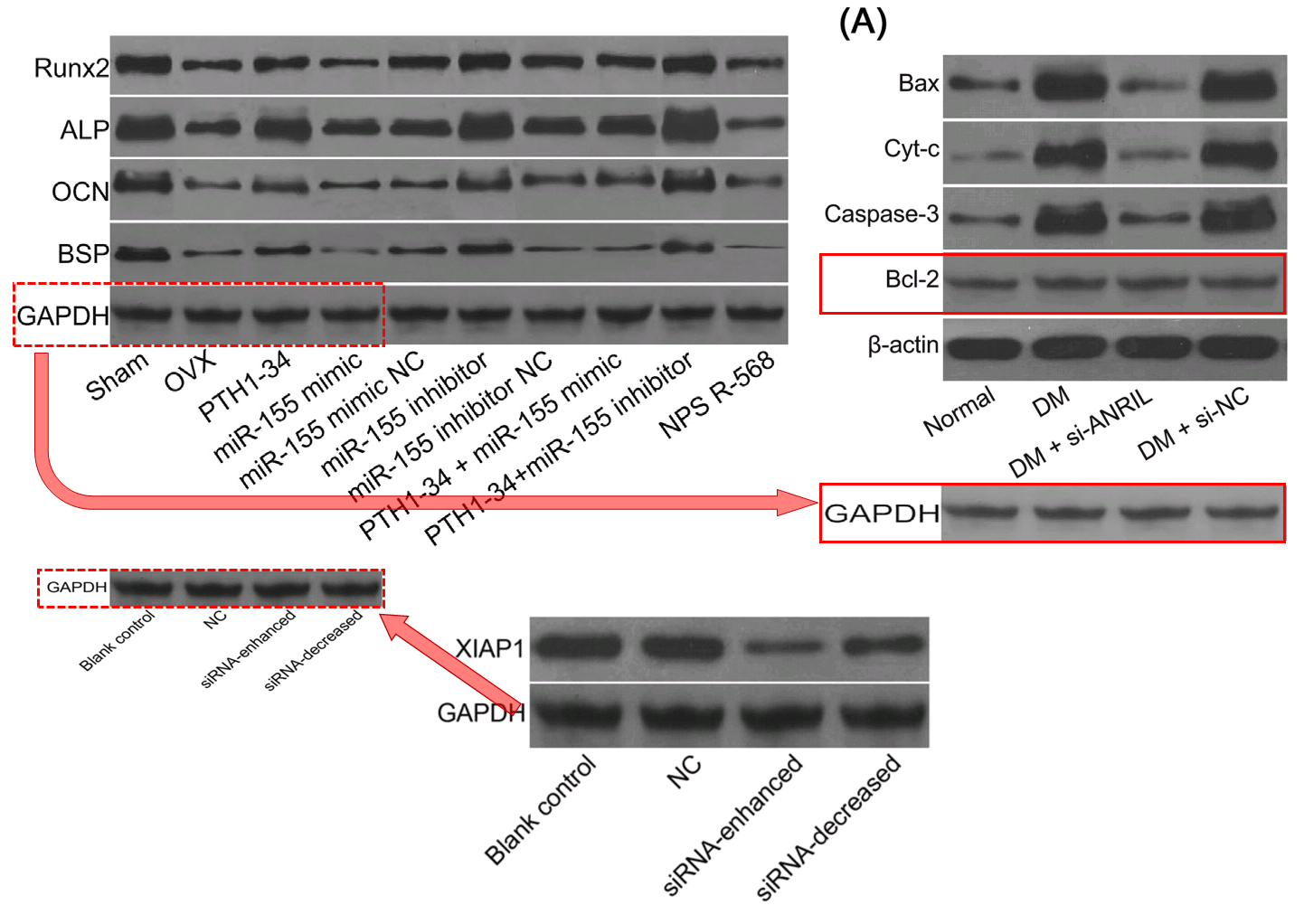
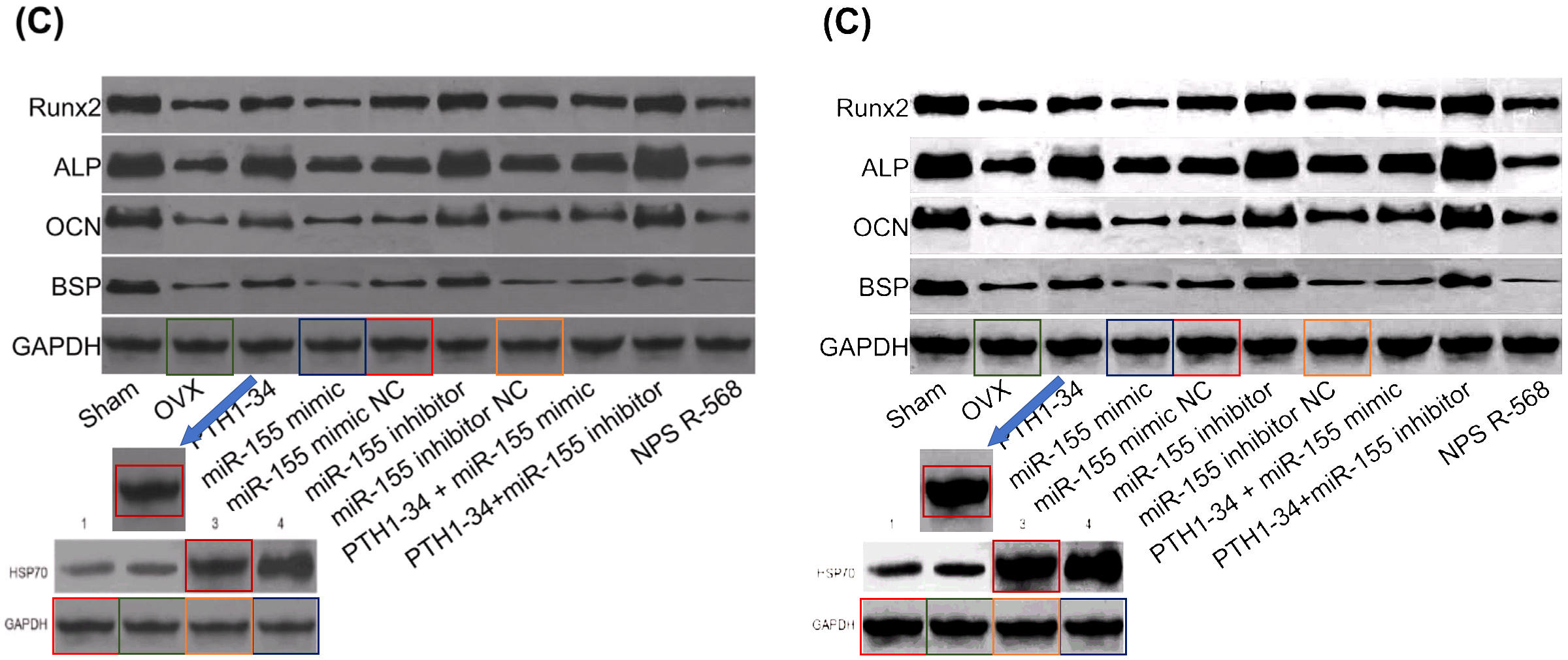
- “In vivo and in vitro effects of PTH1‐34 on osteogenic and adipogenic differentiation of human bone marrow‐derived mesenchymal stem cells through regulating microRNA‐155“. Huan-Sheng Han, Fang Ju, Shuo Geng (2018). Journal of Cellular Biochemistry doi: 10.1002/jcb.26478 [PubPeer].
- “Effects of S100A12 gene silencing on serum levels of anti-inflammatory/pro-inflammatory cytokines in septic rats through the ERK signaling pathway“. Xin Wen, Xin-Rui Han, Yong-Jian Wang, Shao-Hua Fan, Zi-Feng Zhang, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2018). Journal of Cellular Biochemistry doi: 10.1002/jcb.26568 [PubPeer].
- “CUL4B promotes metastasis and proliferation in pancreatic cancer cells by inducing epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway“. Yue-Ming He, Yu-Sha Xiao, Lei Wei, Jia-Qiang Zhang, Cheng-Hong Peng (2018). Journal of Cellular Biochemistry doi: 10.1002/jcb.26643 [PubPeer].
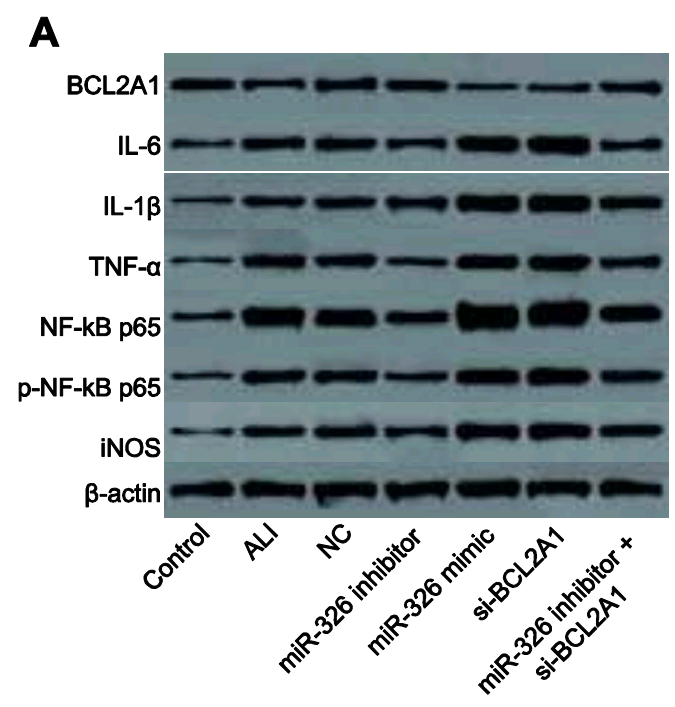
- “Down-regulated long non-coding RNA ANRIL restores the learning and memory abilities and rescues hippocampal pyramidal neurons from apoptosis in streptozotocin-induced diabetic rats via the NF-κB signaling pathway“. Xin Wen, Xin-Rui Han, Yong-Jian Wang, Shan Wang, Min Shen, Zi-Feng Zhang, Shao-Hua Fan, Qun Shan, Liang Wang, Meng-Qiu Li, Bin Hu, Chun-Hui Sun, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2018). Journal of Cellular Biochemistry, doi: 10.1002/jcb.26769 [PubPeer].
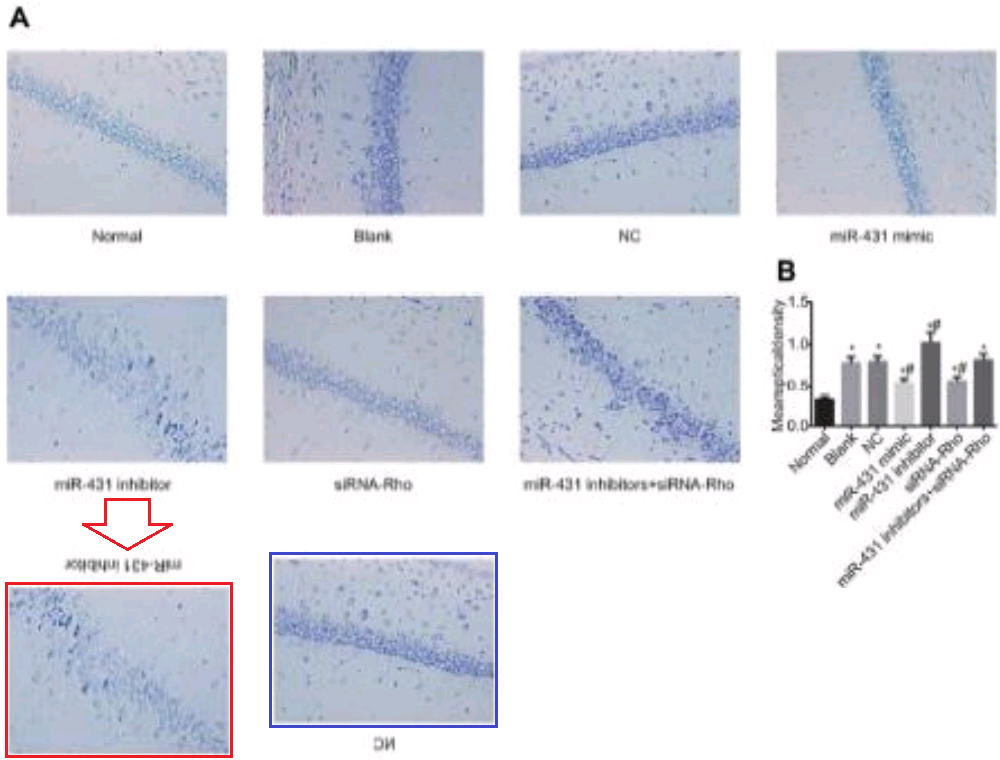
- “MicroRNA-421 suppresses the apoptosis and autophagy of hippocampal neurons in epilepsy mice model by inhibition of the TLR/MYD88 pathway“. Xin Wen, Xin-Rui Han, Yong-Jian Wang, Shan Wang, Min Shen, Zi-Feng Zhang, Shao-Hua Fan, Qun Shan, Liang Wang, Meng-Qiu Li, Bin Hu, Chun-Hui Sun, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2018). Journal of Cellular Physiology doi: 10.1002/jcp.26498 [PubPeer].
- “Effect of microRNA‐186 on oxidative stress injury of neuron by targeting interleukin 2 through the janus kinase‐signal transducer and activator of transcription pathway in a rat model of Alzheimer’s disease”. Dong‐Mei Wu, Xin Wen, Yong‐Jian Wang, Xin‐Rui Han, Shan Wang, Min Shen, Shao‐Hua Fan, Juan Zhuang, Zi‐Feng Zhang, Qun Shan, Meng‐Qiu Li, Bin Hu, Chun‐Hui Sun, Jun Lu, Gui‐Quan Chen, Yuan‐Lin Zheng (2018). Journal of Cellular Physiology doi: 10.1002/jcp.26843 [PubPeer].
- “Suppression of microRNA‐342‐3p increases glutamate transporters and prevents dopaminergic neuron loss through activating the Wnt signaling pathway via p21‐activated kinase 1 in mice with Parkinson’s disease”. Dong‐Mei Wu, Shan Wang, Xin Wen, Xin‐Rui Han, Yong‐Jian Wang, Min Shen, Shao‐Hua Fan, Juan Zhuang, Zi‐Feng Zhang, Qun Shan, Meng‐Qiu Li, Bin Hu, Chun‐Hui Sun, Jun Lu, Gui‐Quan Chen, Yuan‐Lin Zheng (2019). Journal of Cellular Physiology doi: 10.1002/jcp.27577 [PubPeer].
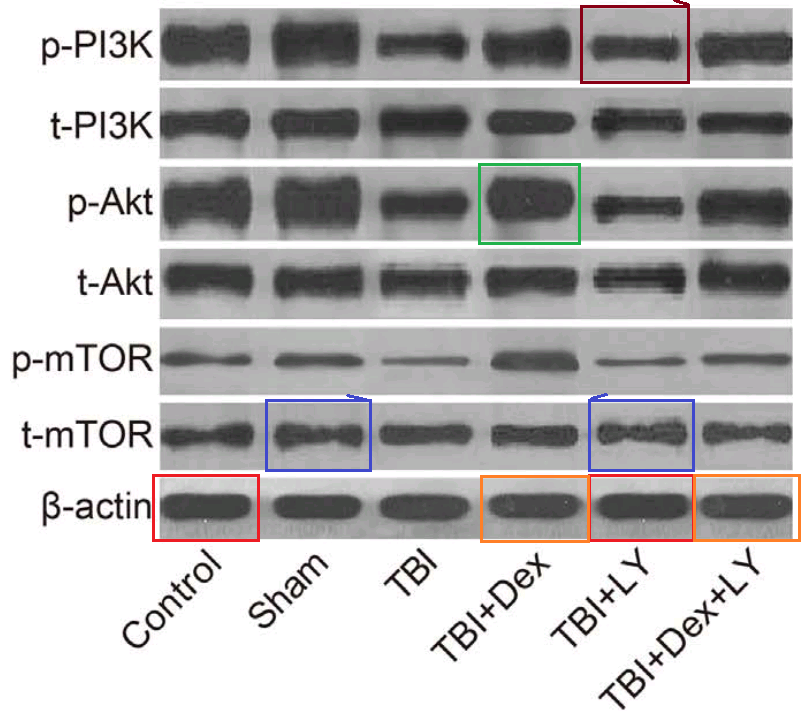
- “Dexmedetomidine exerts neuroprotective effect via the activation of the PI3K/Akt/mTOR signaling pathway in rats with traumatic brain injury”. Min Shen, Shan Wang, Xin Wen, Xin-Rui Han, Yong-Jian Wang, Xiu-Min Zhou, Man-He Zhang, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2017). Biomedicine & Pharmacotherapy, doi: 10.1016/j.biopha.2017.08.125 [PubPeer].
- “MicroRNA-497 promotes proliferation and inhibits apoptosis of cardiomyocytes through the downregulation of Mfn2 in a mouse model of myocardial ischemia-reperfusion injury“. Lei Qin, Wen Yang, Yao-Xin Wang, Zhen-Jun Wang, Chen-Chen Li, Man Li, Jie-Yun Liu (2018). Biomedicine & Pharmacotherapy doi: 10.1016/j.biopha.2018.04.181 [PubPeer].
- “Effects of rehabilitation training on apoptosis of nerve cells and the recovery of neural and motor functions in rats with ischemic stroke through the PI3K/Akt and Nrf2/ARE signaling pathways”. Xiao-Fei Jin, Shan Wang, Min Shen, Xin Wen, Xin-Rui Han, Jun-Chang Wu, Gao-Zhuo Tang, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2017). Brain Research Bulletin doi: 10.1016/j.brainresbull.2017.08.011 [PubPeer].
- “Effect of sevoflurane on the ATPase activity of hippocampal neurons in a rat model of cerebral ischemia-reperfusion injury via the cAMP-PKA signaling pathway”. Tie-Jun Liu, Jin-Cun Zhang, Xiao-Zeng Gao, Zhi-Bin Tan, Jian-Jun Wang, Pan-Pan Zhang, Ai-Bin Cheng, Shu-Bo Zhang (2018). Kaohsiung Journal of Medical Sciences doi: 10.1016/j.kjms.2017.09.004 [PubPeer].
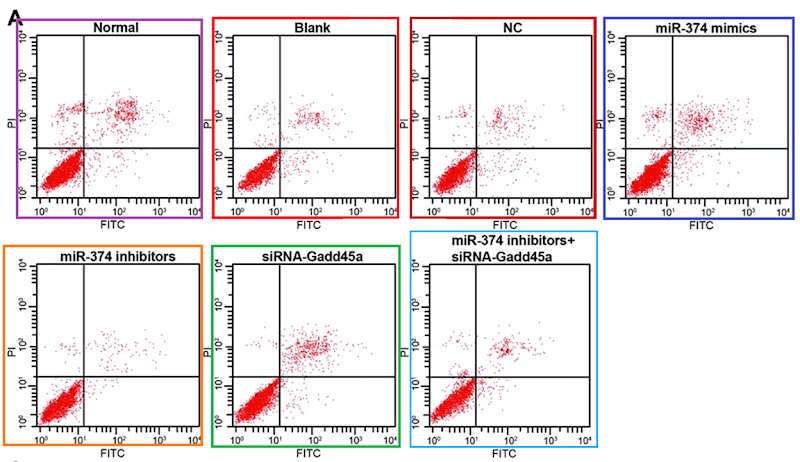
- “Effects of microRNA-374 on proliferation, migration, invasion, and apoptosis of human SCC cells by targeting Gadd45a through P53 signaling pathway“. Xiao-Jing Li, Zhi-Feng Li, Jiu-Jiang Wang, Zhao Han, Zhao Liu, Bao-Guo Liu (2017). Bioscience Reports doi: 10.1042/bsr20170710 [PubPeer].
- “Down-regulation of XIAP enhances the radiosensitivity of esophageal cancer cells in vivo and in vitro”. Xin Wen, Xin-Rui Han, Shao-Hua Fan, Zi-Feng Zhang, Lu Chen, Gao Yi, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2017). Bioscience Reports doi: 10.1042/bsr20170711 [PubPeer].
- “Silencing SUMO2 promotes protection against degradation and apoptosis of nucleus pulposus cells through p53 signaling pathway in intervertebral disc degeneration“. Liu-Zhong Jin, Ji-Shou Lu, Jian-Wen Gao (2018). Bioscience Reports doi: 10.1042/bsr20171523 [PubPeer].
- “MicroRNA-140-5p elevates cerebral protection of dexmedetomidine against hypoxic-ischaemic brain damage via the Wnt/β-catenin signalling pathway”. Xin-Rui Han, Xin Wen, Yong-Jian Wang, Shan Wang, Min Shen, Zi-Feng Zhang, Shao-Hua Fan, Qun Shan, Liang Wang, Meng-Qiu Li, Bin Hu, Chun-Hui Sun, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2018). Journal of Cellular & Molecular Medicine doi: 10.1111/jcmm.13597 [PubPeer].
- “In Vitro Effects of HAS-2 Gene Silencing on the Proliferation and Apoptosis of the MCF-7 Human Breast Cancer Cell Line“. Hong-Yan Zhang, Feng Liang, Fei Wang, Jian-Wei Zhang, Li Wang, Xi-Gang Kang, Jie Wang, Qi-Liu Duan (2016). Cellular Physiology & Biochemistry doi: 10.1159/000453140 [PubPeer].
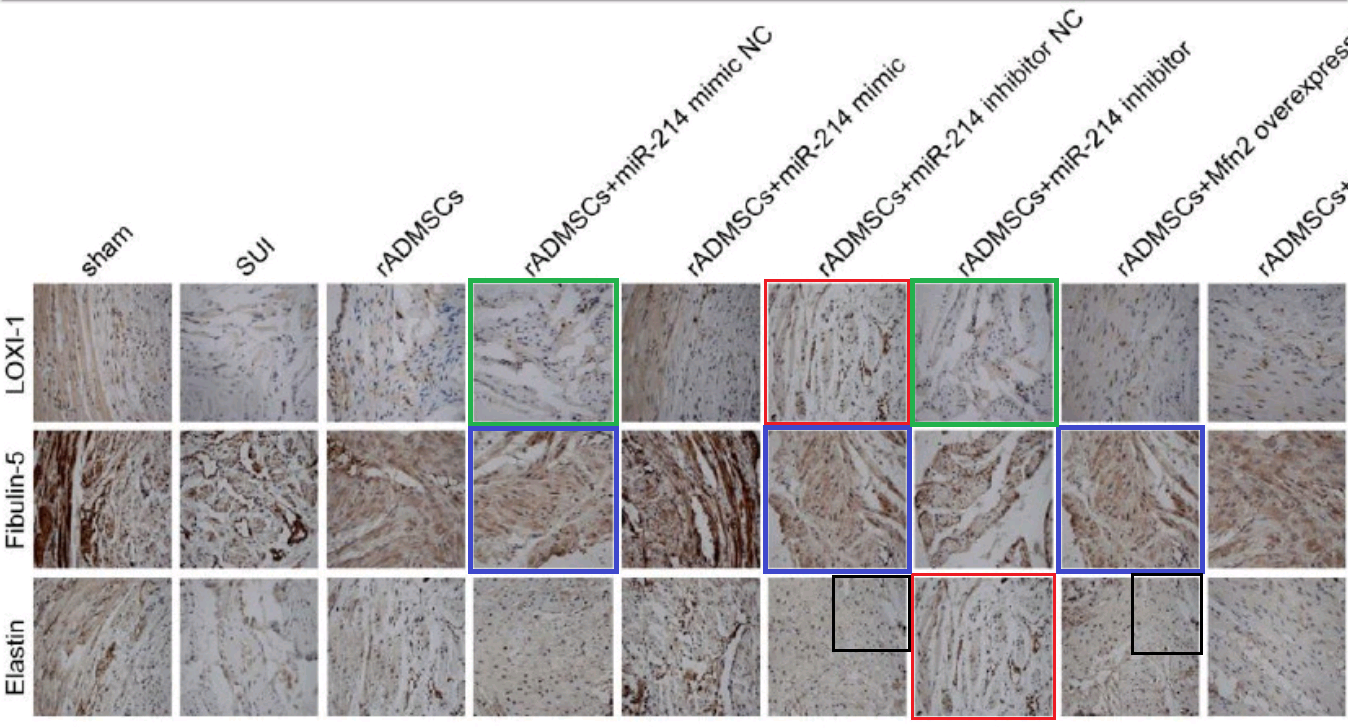
- “MicroRNA-214 Affects Fibroblast Differentiation of Adipose-Derived Mesenchymal Stem Cells by Targeting Mitofusin-2 during Pelvic Floor Dysfunction in SD Rats with Birth Trauma“. Jing Wu, Jiang Li, Wei-Kang Chen, Shang Liu, Jian-He Liu, Jing-Song Zhang, Ke-Wei Fang (2017). Cellular Physiology & Biochemistry doi: 10.1159/000479570 [PubPeer].
- “Effects of MicroRNA-592-5p on Hippocampal Neuron Injury Following Hypoxic-Ischemic Brain Damage in Neonatal Mice – Involvement of PGD2/DP and PTGDR“. Li-Qun Sun, Gong-Liang Guo, Sai Zhang, Li-Li Yang (2018). Cellular Physiology and Biochemistry, doi: 10.1159/000486923 [PubPeer].
- “Effects of MicroRNA-206 on Osteosarcoma Cell Proliferation, Apoptosis, Migration and Invasion by Targeting ANXA2 Through the AKT Signaling Pathway“. Bao-Long Pan, Zong-Wu Tong, Ling Wu, Li Pan, Jun-E Li, You-Guang Huang, Shu-De Li, Shi-Xun Du, Xu-Dong Li (2018). Cellular Physiology & Biochemistry doi: 10.1159/000487567 [PubPeer].
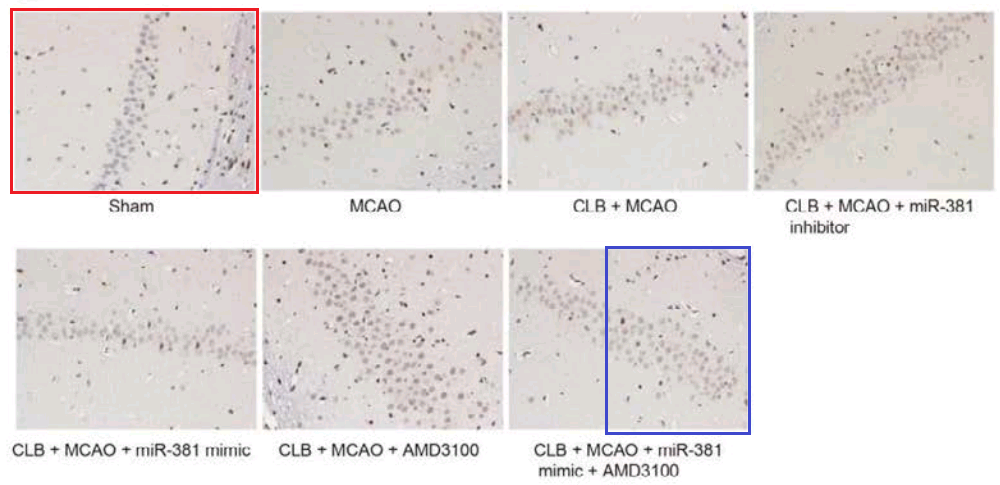
- “MicroRNA-381 Favors Repair of Nerve Injury Through Regulation of the SDF-1/CXCR4 Signaling Pathway via LRRC4 in Acute Cerebral Ischemia after Cerebral Lymphatic Blockage”. Jian-Min Piao, Wei Wu, Zhong-Xi Yang, Yu-Zhen Li, Qi Luo, Jin-Lu Yu (2018). Cellular Physiology & Biochemistry doi: 10.1159/000488821 [PubPeer].
- “Effect of the LncRNA GAS5-MiR-23a-ATG3 Axis in Regulating Autophagy in Patients with Breast Cancer“. Juan Gu, Yueping Wang, Xuedong Wang, Daoping Zhou, Xinguo Wang, Ming Zhou, Zhimin He (2018). Cellular Physiology & Biochemistry doi: 10.1159/000491718 [PubPeer].
- “Suppression of Long Non-Coding RNA LINC00652 Restores Sevoflurane-Induced Cardioprotection Against Myocardial Ischemia-Reperfusion Injury by Targeting GLP-1R Through the cAMP/PKA Pathway in Mice”. Shu-Bo Zhang, Tie-Jun Liu, Guo-Hua Pu, Bao-Yong Li, Xiao-Zeng Gao, Xiao-Liang Han (2018). Cellular Physiology & Biochemistry doi: 10.1159/000493450 [PubPeer].
- “Lentivirus-Mediated Gene Silencing of Lymphocyte Function-Associated Antigen 1 Inhibits Apoptosis of Hippocampal Neurons in Rats with Acute Cerebral Ischemia After Cerebral Lymphatic Blockage“. Jin-Lu Yu, Chao Li, Li-He Che, Ning Xu (2018). Cellular Physiology & Biochemistry doi: 10.1159/000495488 [PubPeer].
- “Inhibition of microRNA-200a Upregulates the Expression of Striatal Dopamine Receptor D2 to Repress Apoptosis of Striatum via the cAMP/PKA Signaling Pathway in Rats with Parkinson’s Disease“. Dong-Mei Wu, Shan Wang, Xin Wen, Xin-Rui Han, Yong-Jian Wang, Min Shen, Shao-Hua Fan, Juan Zhuang, Zi-Feng Zhang, Qun Shan, Meng-Qiu Li, Bin Hu, Chun-Hui Sun, Jun Lu, Yuan-Lin Zheng (2018). Cellular Physiology & Biochemistry, doi: 10.1159/000495649 [PubPeer].
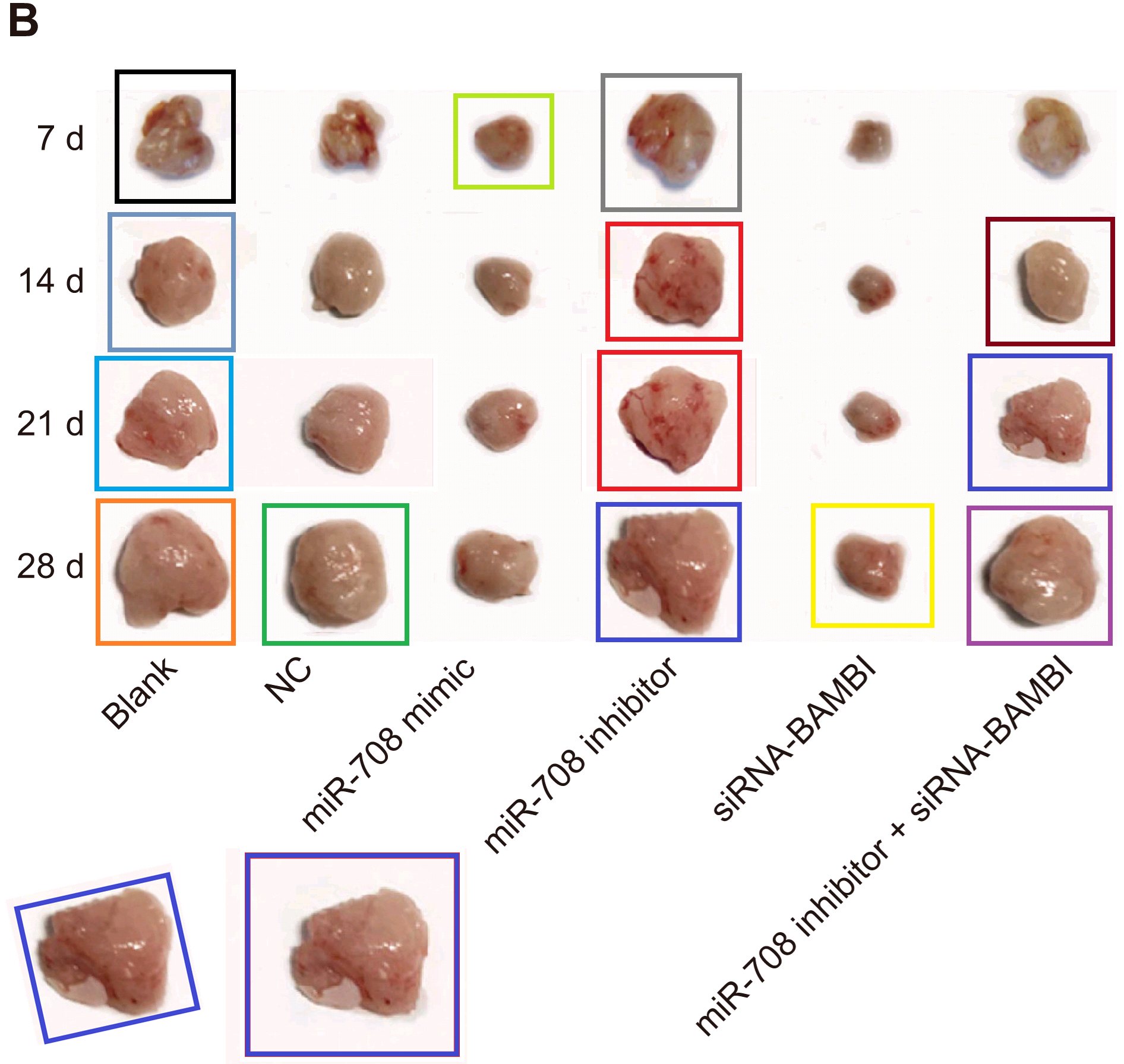
- “Mechanism of MicroRNA-708 Targeting BAMBI in Cell Proliferation, Migration, and Apoptosis in Mice With Melanoma via the Wnt and TGF-β Signaling Pathways”. Hong-Jie Lu, Jing Yan, Pei-Ying Jin, Gui-Hong Zheng, Hai-Lin Zhang, Ming Bai, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2018). Technology in Cancer Research & Treatment doi: 10.1177/1533034618756784 [PubPeer].
- “Effects of CREB1 gene silencing on cognitive dysfunction by mediating PKA-CREB signaling pathway in mice with vascular dementia”. Xin-Rui Han, Xin Wen, Yong-Jian Wang, Shan Wang, Min Shen, Zi-Feng Zhang, Shao-Hua Fan, Qun Shan, Liang Wang, Meng-Qiu Li, Bin Hu, Chun-Hui Sun, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2018). Molecular Medicine doi: 10.1186/s10020-018-0020-y [PubPeer].
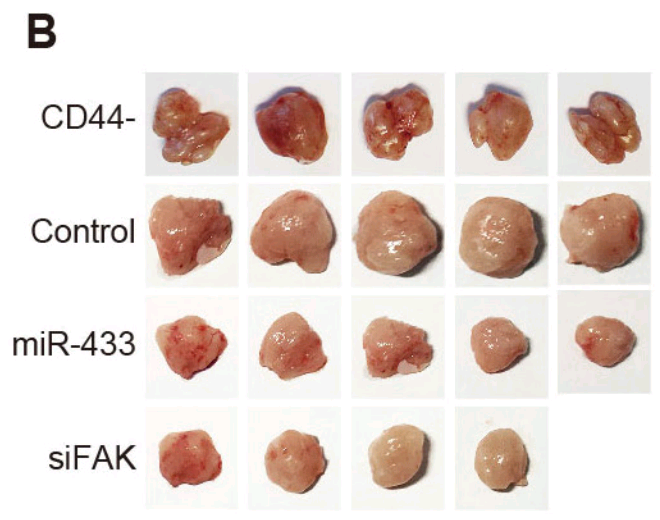
- “MicroRNA-433 inhibits oral squamous cell carcinoma cells by targeting FAK”. Yong-Jian Wang, Zi-Feng Zhang, Shao-Hua Fan, Juan Zhuang, Qun Shan, Xin-Rui Han, Xin Wen, Meng-Qiu Li, Bin Hu, Chun-Hui Sun, Bin Qiao, Qian Tao, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2017). Oncotarget doi: 10.18632/oncotarget.22151 [PubPeer].
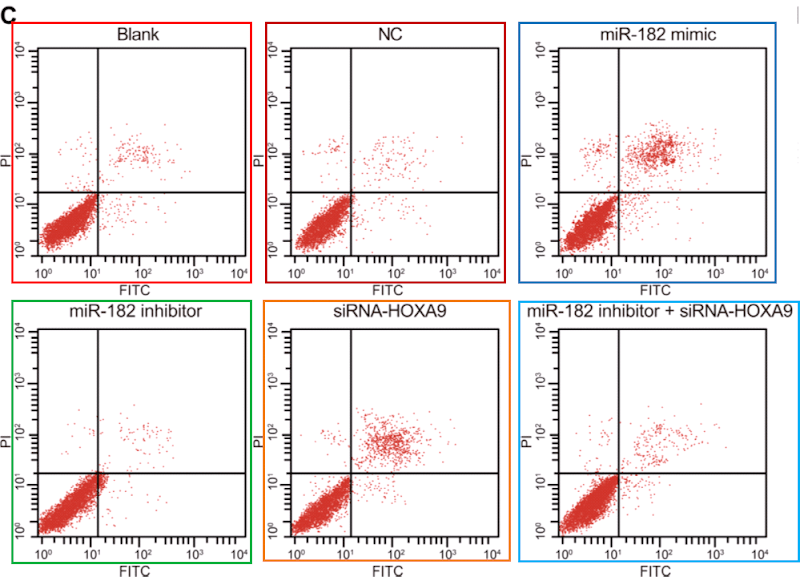
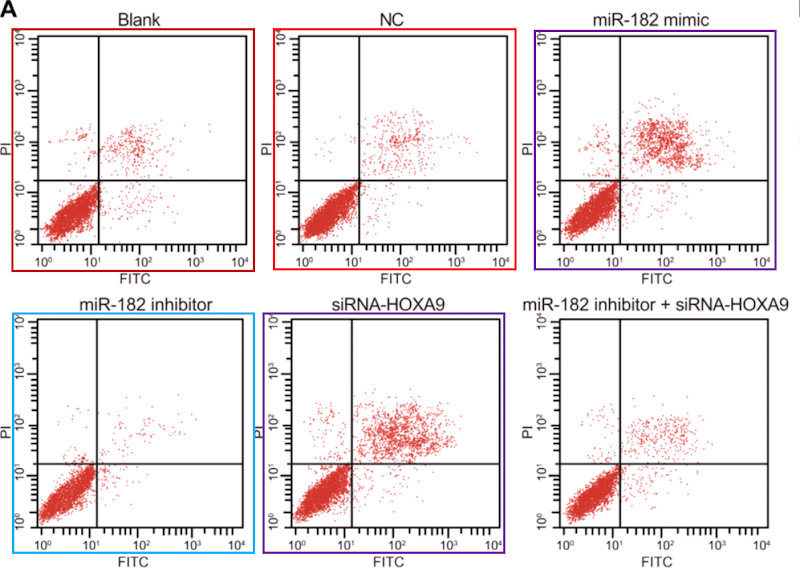
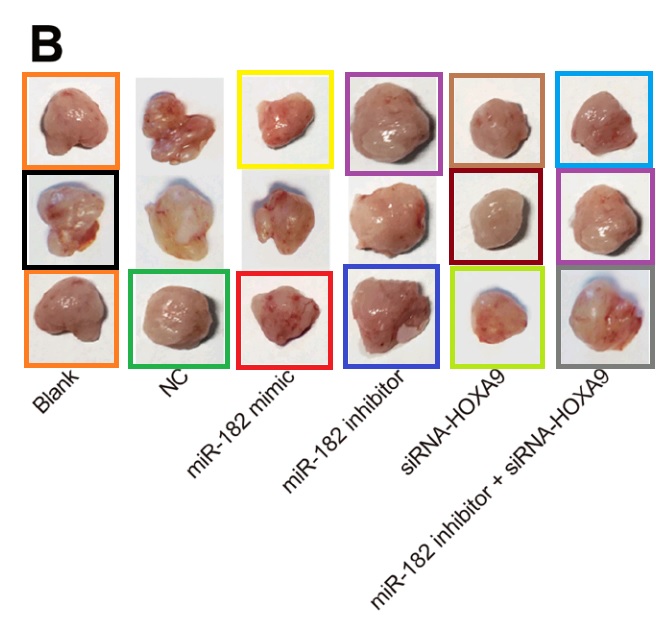
- “MicroRNA-182 downregulates Wnt/β-catenin signaling, inhibits proliferation, and promotes apoptosis in human osteosarcoma cells by targeting HOXA9“. Zi-Feng Zhang, Yong-Jian Wang, Shao-Hua Fan, Shi-Xin Du, Xue-Dong Li, Dong-Mei Wu, Jun Lu, Yuan-Lin Zheng (2017). Oncotarget, doi: 10.18632/oncotarget.21167 [PubPeer].
- “Reduced LINC00540 Expression Inhibits Human Hepatocellular Carcinoma Progression and Metastasis via the NKD2-Dependent Wnt/β-Catenin Pathway”. Dong-Mei Wu, Xin Wen, Xin-Rui Han, Shan Wang, Yong-Jian Wang, Min Shen, Shao-Hua Fan, Zi-Feng Zhang, Qun Shan, Meng-Qiu Li, Bin Hu, Jun Lu, Yuan-Lin Zheng, Gui-Quan Chen (2019). SSRN Electronic Journal, doi: 10.2139/ssrn.3377520 [PubPeer].




China and India go to extreme measures to acquire science and technologies. These two recently wealthy countries take short cuts to acquire projects and ideas instead of discovering their own scientific paths. The former arranges fake conferences and plagerize papers while the latter has become a mega journal producer and printer. Both acquire data before they are published. Gullible scientists attend fake conference in exotic locations paying their own money. No one accounts for how much of NIH money goes in travel to these fake conferences by funded scientists.
LikeLike
IMO, there may not be an uncontainable, all enveloping cancer that will kill western science, as some fear. There is hope, as I posit here: If these authors with fake papers can just cite other authors of other fake papers very frequently, we might be able to create a self contained system that generates crap separate from the reproducible material. Hmmm: self citation of fake papers–>Chinese authors get more Chinese govt grant money–> authors pay for fake papers—>fake papers get published in once-respectable western journals now contaminated with crap (eg Mol Imm) –>fake papers cite each other, and the cycle continues. The “energy source” is Chinese Govt money, and the waste products (analogous to CO2 and H2O) are crap papers. I in all modesty want to call this the NMH cycle, because I thought of this first. Self contained crap, and sequestered from quality science. Now if only the powers in academia can call this out without being shamed by social justice PC types.
LikeLike
“It would be helpful if Acknowledgements sections also included a note of which papermill had provided the manuscript, or which consultants the authors had paid to adorn their own manuscript with additional figures and results.” This is a key point. No matter how much we go on about perverse incentives, the authors of these papers reveal a total lack of both scientific morality and competence when they publish the work of others as their own. That is grounds for summary dismissal where I work, and it should be at every genuine research institute.
It would also be useful if journals would keep track of the reviewers who fall for those stupid megablots and cookie-cutter flow diagrams, and who don’t seem to know their hearts from their brains, livers and nether bits.
LikeLike
You mean like Carlo Croce? I realize I may be sued for this comment.
LikeLiked by 1 person
Not likely. I hear he already ran up a big lawyerin’ bill that no honest man can pay.
LikeLike
Ah, but Croce can sell his highly valuable and original Baroque paintings! Although Smut Clyde has a compelling theory that these old masters are just as fake as Croce’s cancer research.
LikeLike
Are you crazy, Owlbert? Sack all fraudsters? Who will provide all the papers then? People like you are not productive enough.
LikeLike
That’s true. A few hundred bottom-feeder papermill journals will have to go if only real scientists are left. And Nature as well, I guess. No loss.
LikeLike
Pingback: Propaganda - Ocasapiens - Blog - Repubblica.it
NIH seems to be coming up with some solutions, https://nexus.od.nih.gov/all/2020/07/08/addressing-foreign-interference-and-associated-risks-to-the-integrity-of-biomedical-research-and-how-you-can-help/
It would be interesting to see any of these authors have been visiting students in or postdocs in USA and worked in NIH funded labs.
LikeLike
There is a solution that nobody seems to consider: stop rewarding publications, ban perverse incentives and reconsider mandatory use of a foreign language. It is beyond unethical to force foreign authors to publish in English. And why expect clinicians to be tech savvy and produce microscopic images without using the services of third parties? The root cause of this problem is a hidden conflict of interest wbich is motivated by the continuous need to feed publishers products
LikeLike
“It is beyond unethical to force foreign authors to publish in English.” They are not forced to do so.
LikeLike
Isn’t publishing in top tier journals a mandatory requirement for tenure?
LikeLike
“The Acknowledgements in [13] thank the research subjects for their selfless commitment to Science. Laboratory rats so seldom receive the credit they deserve for their sacrifices. “Acknowledgments: We would also like to thank all participants enrolled in the present study.”
The Acknowledgements in https://ese.arphahub.com/article/52201/ lack these details. Are the authors of [13] willing to transfer their text to https://ese.arphahub.com/article/52201/ ?
LikeLike
It is absurd to say that “It is beyond unethical to force foreign authors to publish in English.” There are many other languages of publishing, and I often see that Chinese authors publish the same thing twice: once in a Chinese journal what the 70% of the World cannot read, and also in English, what 99% of the World can read (besides the Authors). Anyway, today English is the universal language of science, and many other sectors of life. But there are people who always pick up randomly things to feel offended about, so look forward the next riots to ban English in Science.
LikeLike
“I often see that Chinese authors publish the same thing twice: once in a Chinese journal …”
Interesting. Do authors of identical publications use identical duplicate and fake images in their Chinese publications? Has anyone compared a Chinese article to its English version? Red flags here, perhaps the issue is a unique phenomenon!!!
LikeLike
I know that in the communist era in the Eastern block it was normal to publish a work in a national journal, and afterwards in a Russian one, or if you could manage to sneak the manuscript to the West, publish it in English.
LikeLike
Pingback: Dead horses won’t flog themselves – For Better Science
Pingback: Victims as perpetrators – For Better Science
A recent tranche of retractions of ‘contractor’ papers from Bioscience Reports included “Effects of microRNA-146a on the proliferation and apoptosis of human osteoarthritis chondrocytes by targeting TRAF6 through the NF-κB signalling pathway”. It’s a whole three-act play.
Act 1. July 12 2017: Published, with admiring commentary by C.West and M. F. McDermott, urging readers to consider the implications of this stimulating, ground-breaking research with its “compelling data”.
Act 2. September 28 2020: Expression of Concern.
Act 3. June 8 2021: “This Retraction follows an Expression of Concern relating to this article previously published by Portland Press.”
It doesn’t quite reach the heights of Ibsen or Chekov.
LikeLike
The Yuan-Lin Zheng & Dong-Mei Wu case received formal investigation by NSFC and here is the results:
https://www.nsfc.gov.cn/publish/portal0/tab442/info85495.htm
Dong-mei Wu is banned for life from applying grant from NSFC. This is rare but she totally deserved that. Unfortunately, Yuan-Lin Zheng and Jun Lu did not received as harsh punishment as Wu got. My guess is that they both are in the leadership of the university.
LikeLike
Pingback: The Rise of the Papermills – For Better Science
Pingback: The papermills of my mind – For Better Science