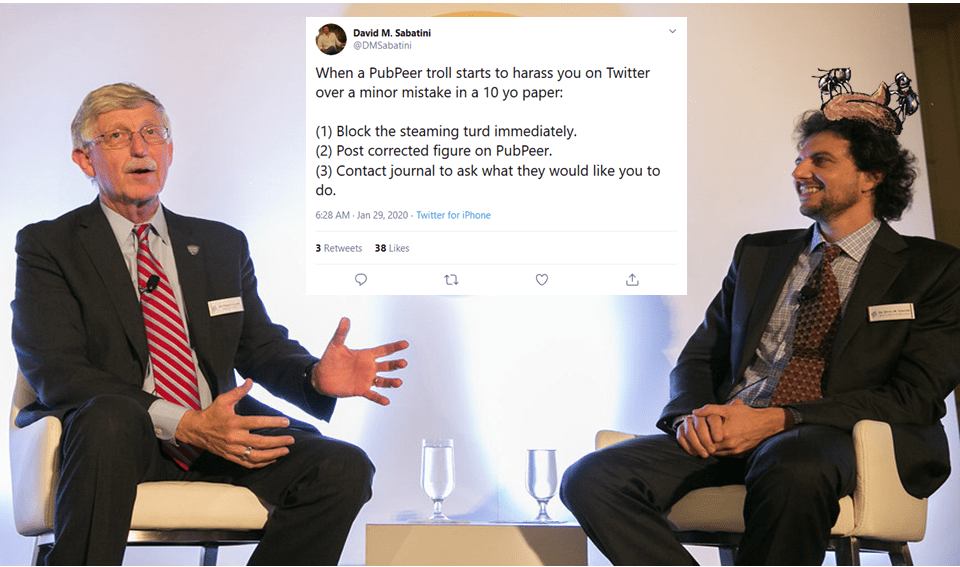
“PubPeer may have had a role at some point, but as far as I can tell, is now a platform for resentful, anonymous, petty, failed scientists to harass those who actually make discoveries“
Professor David M Sabatini, mTORman
David Sabatini is MIT professor and a titan of molecular biology, an absolute superstar or actually, a superhero of science. The Whitehead Institute principal investigator calls himself mTORman, because he is credited with the discovery of the mTOR kinase, which regulates many cellular processes and is therefore an invaluable target for the pharma industry.

mTORman’s chief superhero power is apparently to be able to certify all conclusions of his top-rank papers as unaffected, despite image duplications therein, and without ever looking into raw data. Instead, Sabatini sends his first authors to replace the offending figures and to warn everyone not to question their papers. And those who insist he checks the data consistency anyway are described as “steaming turds”. Those like Strobilanthes Asper who recently raised concerns about data integrity in Sabatini’s papers, get off lightly: they are merely “failed scientists”. Apparently, these days this is the dictionary definition of
Scientist, successful: a person employed at elite research institution, able to regularly place papers in journals with highest impact factor despite data irregularities and unverifiable research results. Is generally above scrutiny from journals and universities; instils the fear of God in critics.
Some time ago, Sabatini’s science was mildly criticised on PubPeer, and back then no concerns of data integrity were raised. The great man was not amused. The Harvard superstar, who gets angry easily and throws expletives at his critics, protested the anonymity of a PubPeer commenter. After all, a man is entitled to have revenge, and not knowing your critic’s identity is not helpful here.
I think Strobilanthes Asper did a great job flagging Sabatini’s papers on PubPeer and would like to open with a particularly disastrous one. The MIT superstar is penultimate author there, meaning his lab contributed the second-biggest share to that paper. The last author is Dos Sorbassov, former mentee of Sabatini, who then moved to MD Anderson in Texas (a notorious place), and who is now back in his home country Kazakhstan as professor at the Nazarbayev University (named after the country’s undead post-Soviet long-term dictator).
D Boulbes, CH Chen, T Shaikenov, NK Agarwal, TR Peterson, TA Addona, H Keshishian, SA Carr, MA Magnuson, DM Sabatini , DD. Sarbassov Rictor phosphorylation on the Thr-1135 site does not require mammalian target of rapamycin complex 2 Molecular cancer research : MCR (2010) doi: 10.1158/1541-7786.mcr-09-0409
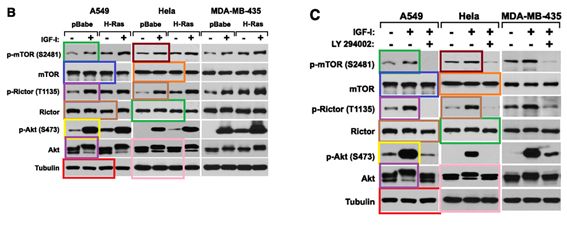
Horrible, right? An orgy of Photoshop fraud, a retraction is too mild for this awful paper. So far, mTORman saw no reason to jump to action.
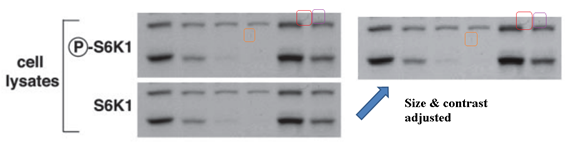
This makes you suspicious of another Sarbassov & Sabatini JBC 2005 cooperation, where a gel for total protein looks exactly like the phosphorylated one (they are supposed to look somewhat similar in band shapes, but never identical). Or, is this really an innocent mistake of oversight, spotted by another PubPeer commenter in an old Science paper from Sabatini lab:
D D Sarbassov, DA Guertin, SM Ali, DM Sabatini Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex Science (2005) doi: 10.1126/science.1106148
Again, S6K1 blots are accidentally duplicated. As if there was some kind of bug with the S6K1 data? Sarbassov’s papers are truly worth studying, more evidence is listed in the comments below this article. To be fair, Sabatini reacted immediately after I pointed him to his Boulbes et al 2010 paper and the next one, in Nature: he blocked me on Twitter and described me as a “steaming turd”. Which I presume also precludes any possible inquiries at Whitehead and MIT?
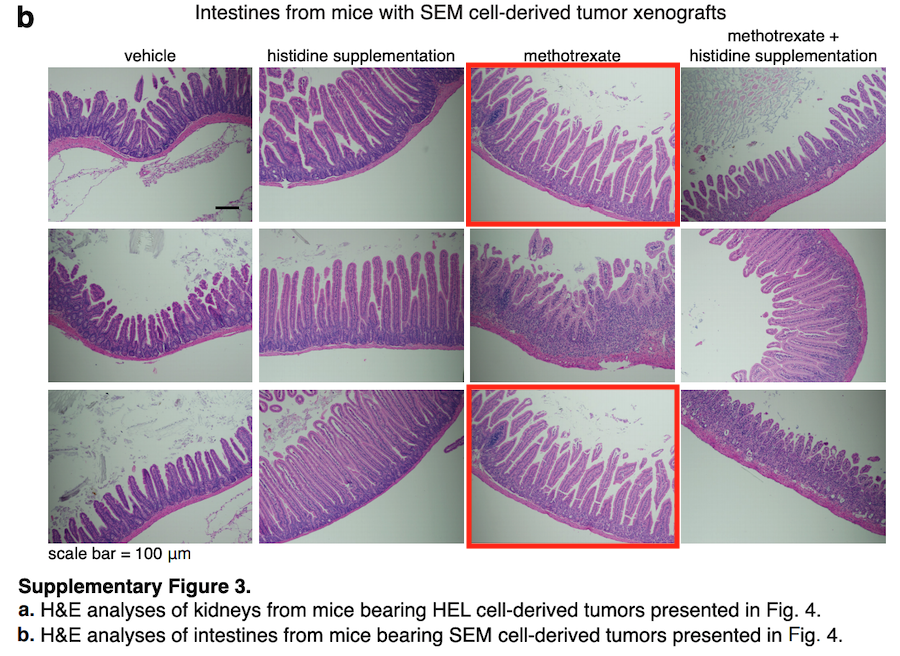
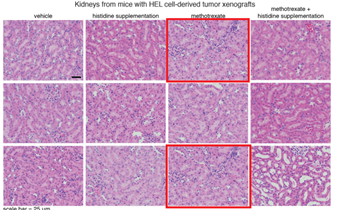
N Kanarek , HR. Keys , JR. Cantor , CA. Lewis , SH Chan , T Kunchok , M Abu-Remaileh , E Freinkman , LD. Schweitzer , DM. Sabatini Histidine catabolism is a major determinant of methotrexate sensitivity Nature (2018) doi: 10.1038/s41586-018-0316-7
Three image duplications inside same Figure 11, a and b. A rather new paper, why did nobody notice that? Or was Nature afraid to tell Sabatini, fearing he will publish with Cell again next?
The first author Naama Kanarek, now group leader in Harvard, sees it all very relaxed:
“we will also ask Nature if they wish to correct it on the online version of the paper. We emphasize that this mistake does not change the conclusion of the figure or any of the statements made in the paper.“
When Harvard and MIT folks keep telling you such things, it does sound a bit like a threat. The paper is not even 2 years old, and they already warn you not to ask for a correction, apparently exactly because this is in Nature. whom they also expect to do as told. Can someone tell our scientific elites it is not for them and their editor buddies to decide when their shoddy attitude to data presentation affects the conclusions, but for the scientific community? Especially since they refuse to examine the raw data or to test the reproducibility of their results, as Sabatini made clear on Twitter?
How does one recover old data stored somewhere in Sabatini lab via email discussions with first author who moved on to open a lab of their own? No wonder the hastily procured replacements do not seem to exactly fit. Here is the next figure for Dr Kanarek of Harvard to fix, in that same Nature 2018 paper:
Turns out, the quantifications presented in Figure 2 are based on copy-pasted numbers, as evidenced by the raw data authors had to supply according to new Nature guidelines. Does this affect any of the conclusions, Dr Kanarek?


Kanarek had replacement data ready and explained:
“In my attempt to make the source data files clear I transposed each of the data sets and named the sets by cell line, treatment and metabolite. It was a mistake because it caused multiple errors.“
Nada Y. Kalaany , David M. Sabatini Tumours with PI3K activation are resistant to dietary restriction Nature (2009) doi: 10.1038/nature07782

The coauthor Nada Kalaany, now associate professor at Dana Farber / Harvard Cancer Institute replied on PubPeer and announced a possible correction:
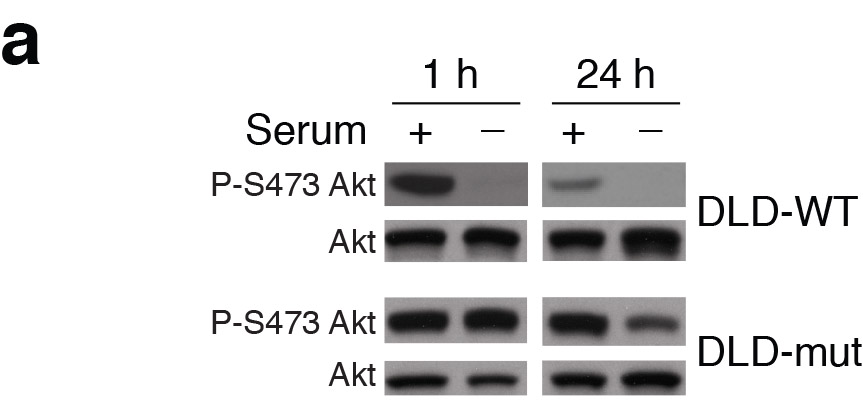
“This error does not impact the conclusions of the paper. Nevertheless, we will contact the Nature Editorial Office to let them know about this mistake and its correction.“
Where the replacement gel exactly came from is not clear, the new phospho-AKT band “smiles” upwards, while the total AKT band “smiles” downwards, a rare thing to happen were it the same gel re-probed. The new replacement blot also has a different colouring, as another PubPeer commenter noticed. All of which lets one wonder where exactly Dr Kalaany got it from.
In the next instance, a confocal microscopy image was reused in a different case first in a bioRxiv preprint, and then in a PNAS paper. The experimental settings were very different. Sabatini was penultimate, Jared Rutter of University of Utah last author.
CK Kikani , X Wu , S Fogarty , SA Woo Kang , N Dephoure , SP Gygi, DM. Sabatini , J Rutter Activation of PASK by mTORC1 is required for the onset of the terminal differentiation program Proceedings of the National Academy of Sciences (2019) doi: 10.1073/pnas.1804013116
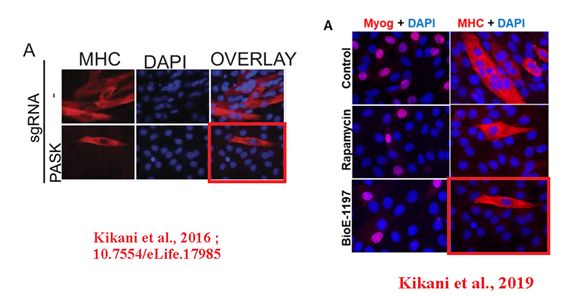
First author Chitnan Kikani is now assistant professor at University of Kentucky, and he announced to fix both the preprint and the PNAS paper:
“we wish to point out that our erroneous insertion of a panel does not aid or affect data interpretation in any way. […] We are now pursuing the submission of an erratum, wherein we will explain the error and replace the image with the appropriate one.“
The next duplication is also puzzling.
J Wedel , S Bruneau , K Liu , SW Kong , PT. Sage , DM. Sabatini , M Laplante, DM. Briscoe DEPTOR modulates activation responses in CD4 + T cells and enhances immunoregulation following transplantation American journal of transplantation (2019) doi: 10.1111/ajt.14995

The first author Johannes Wedel, originally from Germany, is faculty member in the Harvard lab of David Briscoe, who is last author of that paper. Wedel explained on PubPeer, “on behalf of all authors”:
“this minor error and its correction (see below) does not affect the summary of n=8 mice/group illustrated in the same Figure (Fig.1F). This likely resulted from a copy and paste issue during the preparation of the figure and went unnoticed during the submission, review and publication of the manuscript. Also, importantly this minor error and its correction does not change the interpretation of data or conclusions drawn from the extensive additional analyses (including full transcriptomic data, in vitro and in vivo functional analyses) illustrated in the other figures. Nevertheless, we have notified the Editorial Office of the American Journal of Transplantation about this error and its correction.“
Maybe not so fast, Herr Doktor Wedel. Surely everyone can accidentally reuse a FACS dataset, but: Since the quantifications are different (11.9 vs 18.0), the most benevolent explanation would be that someone was routinely analysing flow cytometry samples of the same experiment with different gate settings.
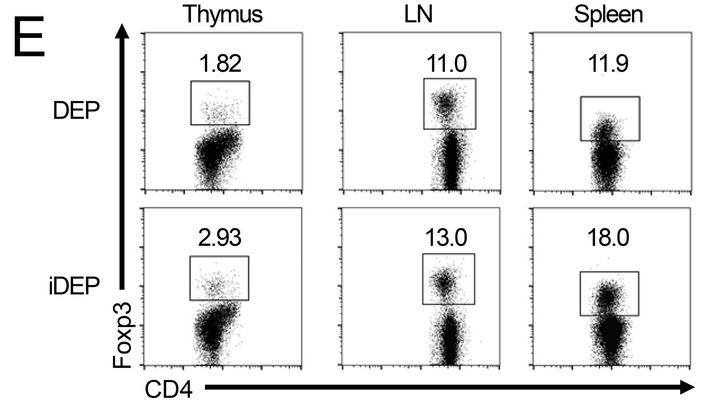
A very bad and inappropriate practice (even if other German experts see it differently). Miraculously, Wedel’s replacement figure had the same quantification number 11.9, now based on Ersatz flow cytometry plot (right).
It is not reassuring that neither Briscoe nor Sabatini nor other elite labs have a problem with such approach to flow cytometry. On the other hand, it does produce the “right” results for the right journals. The Journal of Biological Chemistry (JBC) is however the wrong journal to put bent data in.
Q Liu , S Kirubakaran , W Hur , M Niepel , K Westover, CC. Thoreen, J Wang , J Ni , MP. Patricelli , K Vogel , S Riddle , DL. Waller , R Traynor , T Sanda , Z Zhao , SA. Kang , J Zhao , AT Look, PK. Sorger , DM. Sabatini , NS. Gray Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics Journal of Biological Chemistry (2012) doi: 10.1074/jbc.m111.304485
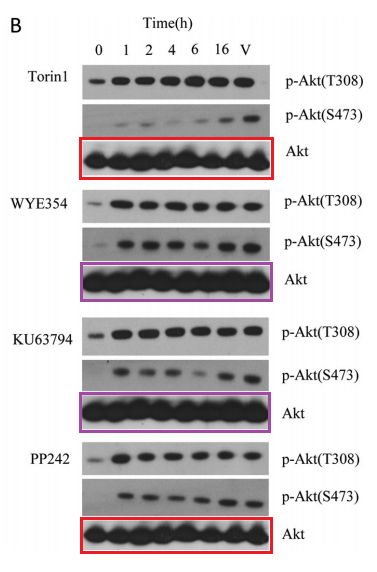
That figure was originally flagged for the re-used Akt western blots, which function as loading control here. A mistake of oversight maybe, until someone else commented:
“Akt panels appear to show 8 lanes as a control for a 7 lane blot“
Oops. This suggest that the authors possibly did not have the correct Akt blots at hand in the first place, and used some unrelated library loading controls, copy-pasted two times each, without even bothering if they at least show the same number of samples.
But how do we know the blots were equally loaded then? Simple: because the last authors are Sabatini and Nathaniel Gray of Dana Farber Cancer Institute in Harvard. QED. This is also why the first author of another Sabatini-Gray co-production Carson Thoreen (now associate professor at not just somewhere, but at Yale) replaced a western blot loading control in Thoreen et al JBC 2009 while warning:
“We would like to emphasize that these changes have no impact on any of our original conclusions.“
Another commenter pointed out that “the background color of this new blot is slight different than all the other blots (is more blueish, while the others are gray)“, which lets one wonder again where the resourceful first author got it from. If it was the original, correct scan from 2009, it would have been the same colour as the rest of the figure, no?
Yale professor Thoreen would probably also say none of that affect the conclusions of this paper of his:
Y Sancak , CC. Thoreen , TR. Peterson , RA. Lindquist , SA. Kang , E Spooner , S A. Carr, M. Sabatini PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase Molecular Cell (2007) doi: 10.1016/j.molcel.2007.03.003
First author Yasemin Sancak is now assistant professor at University of Washington. What is it with that wretched ribosomal protein S6 kinase S6K which always needed some tinkering with?
This other Sabatini collaboration is another chapter in 50 shades of Gray, very painful and excruciating to watch:
Q Liu , C Xu , S Kirubakaran , X Zhang , W Hur , Y Liu , NP Kwiatkowski , J Wang , KD Westover , P Gao , D Ercan , M Niepel , CC Thoreen , SA Kang , M P Patricelli , Y Wang , T Tupper , A Altabef , H Kawamura , KD Held , DM Chou, SJ Elledge, PA Janne, KK Wong, DM Sabatini, NS Gray Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR Cancer Research (2013) doi: 10.1158/0008-5472.can-12-1702
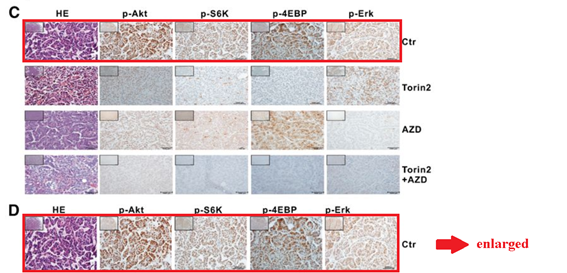
Somehow the histological sections for animals sacrificed after 2 days of treatment (Figure 5C) are identical to those sacrificed after 4 weeks (Figure 5D), only stretched. A mistake? Sure it is, just like these duplications are, once again that S6K protein misbehaved:
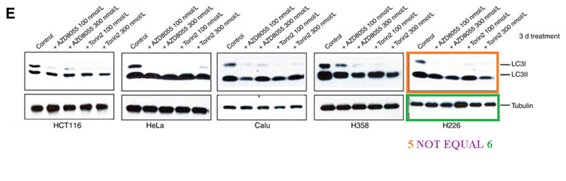
The two papers are merely 8 years old, but so far the authors remained silent. There is little incentive for Gray to bother with PubPeer criticisms in the first place, because the director of his Ludwig Center at Dana Farber is none other by Bob Weinberg, who is a god in his own right and as such above such things.
Speaking of gods. Before forming an opinion about this Cell paper from the lab of David Root at Broad Institute, you should ask yourself: are you worthy? It features the cancer research god Bill Hahn and the Broad Institute director Eric Lander, next to Sabatini. On your knees, now.
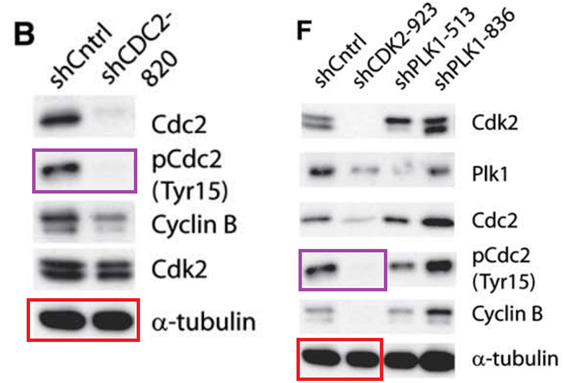
J Moffat, DA. Grueneberg, X Yang, SY Kim, AM Kloepfer, G Hinkle, B Piqani, TM Eisenhaure, B Luo, K. Grenier, AE Carpenter, SY Foo, SA. Stewart, BR. Stockwell, N Hacohen, WC Hahn, ES Lander, DM Sabatini, DE Root A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen Cell (2006) doi: 10.1016/j.cell.2006.01.040
The first author Jason Moffat, now professor at University of Toronto, replied:
“We are checking into this.”
Moffat did not say whether the conclusions are affected or not, but in any case: only a steaming turd of a failed scientist would doubt his Cell paper now.
The next paper has a minor duplication of a FACS plot, but the Korean first author Joon-Ho Sheen is presumably too busy to reply, being a senior scientist at LG.
JH Sheen , R Zoncu , D Kim , DM. Sabatini Defective Regulation of Autophagy upon Leucine Deprivation Reveals a Targetable Liability of Human Melanoma Cells In Vitro and In Vivo Cancer Cell (2011) doi: 10.1016/j.ccr.2011.03.012
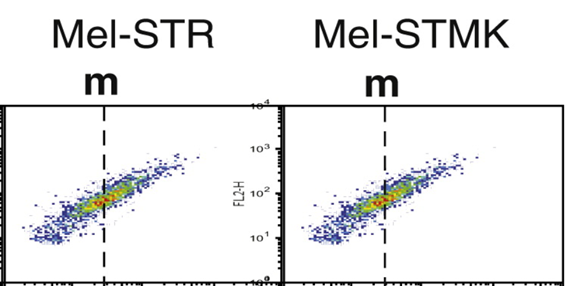
Sabatini, in whose lab the original data remained, cannot be bothered. In 2021 that paper will be 10 years old and it will be inappropriate to ask him to check for any eventual raw data anyway.
Also here nobody bothered to reply. The first author Liron Bar-Peled is now assistant professor and principal investigator at Dana Farber/Harvard Cancer Center.
L Bar-Peled, LD. Schweitzer, R Zoncu, David M. Sabatini Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1 Cell (2012) doi: 10.1016/j.cell.2012.07.032
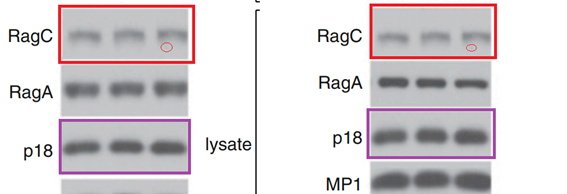
This time it does not look like a mistake of oversight. The Figures 1A and 1B contain 12 western blots each, labelled as clearly different assays with different protein constructs. And yet the RagC and p18 panels were reused, and stretched, which made them look dissimilar.
There are other issues with Sabatini’s publication record: a collaborative paper with a Chinese group contains an unashamedly fake western blot, or a shady paper with the notorious Italian cheater duo Carmine Settembre and Andrea Ballabio (Settembre et al EMBO J 2012).
More recently, concerns were raised that the authors misinformed peer reviewers and readers about the specificity of the NEK10 antibody central to the paper’s main findings: the paper Chuvukula et al Nature Medicine 2020 is brand new. Similar concerns were noted for a slightly younger Nature paper from Sabatini lab, Wolfson et al Nature 2017. But the mighty mTORman already made clear on Twitter that he does not intend his papers for the scientific community, only for the exclusive readership of editors at elite journals. Whether or not the presented scientific results are any reliable, does not in any way affect the perfectly factual and reproducible conclusions that those were published in Nature and Cell.
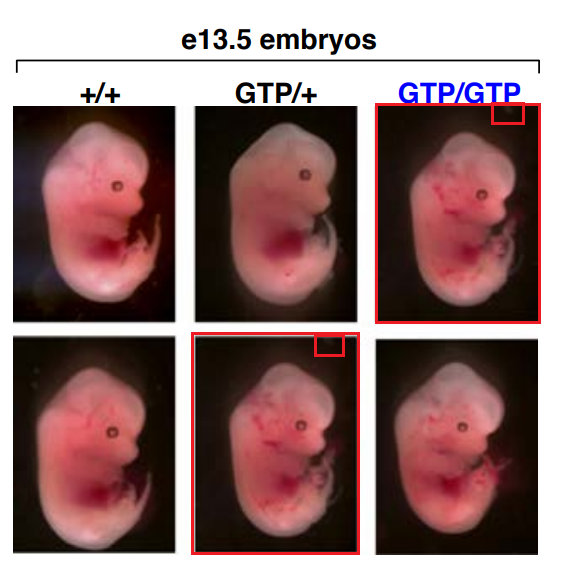
There is also a duplicated picture of a mouse embryo in Sabatini’s own paper in Nature, Efeyan et al 2013, co-authored by his dad, David Sabatini Sr. There, the first author Alejo Efeyan, now group leader at CNIO in Madrid, informs everyone:
“It is important to note that neither this very minor mistake nor its correction affect the validity of the data, its interpretation, conclusions drawn, or any other aspect of the published paper.“
You steaming turds have been warned.

This article was updated on 30 and 31 January 2020.
Update 21.08.2021
Sabatini was sacked after an investigation found that he “violated the Institute’s policies on sexual harassment among other Whitehead policies unrelated to research misconduct”.
An email by Whitehead Institute director Ruth Lehmann was shared with the employees:
I am writing to let you know that David Sabatini, a member of Whitehead Institute, is no longer associated with either the Whitehead Institute or the Howard Hughes Medical Institute, effective immediately.
Dr. Sabatini’s departure comes on the heels of his receipt of a report laying out the findings of an independent investigation into the culture and working environment of his lab. This investigation was precipitated by a Diversity, Equity and Inclusion survey commissioned and conducted last winter which collected data and comments on the culture across the Institute. The results of this survey identified issues of particular concern in the Sabatini Lab and that led to the appointment of Hinkley Allen & Snyder LLP to investigate the Sabatini Lab. In sum, the investigation found that Dr. Sabatini violated the Institute’s policies on sexual harassment among other Whitehead policies unrelated to research misconduct.
Dr. Sabatini’s departure has significant implications for the 39 members of his lab, four of whom are HHMI employees; the remainder are Whitehead employees. Whitehead human resources personnel will be conducting one-on-one meetings with all 39 – next week – to help effectuate a plan to ensure their smooth transition to another lab setting so that they may continue their work in pursuit of their career goals.
I am and will always be steadfast in my commitment to providing an inclusive, supportive environment for the training and research of our community.Ruth Lehmann
Maybe the misconduct investigation set off other complaints. Pier Paolo Pandolfi was also sacked in Harvard for sexual harassment unrelated to research misconduct.
That’s what we steaming turds achieved.
For more mTOR fraud, read here:

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism will be most appreciated!
€5.00






I don’t think Sabitini appreciates that often the reason “failed” scientists get created is because of fraud going on in big-shot labs. Fraud is great at creating papers, lots of papers get grant money and promotions for the person in charge. Getting grants is extremely competitive, so labs that create crap papers get the grant money, while honest labs do not, rendering a tenured honest faculty as deadwood (which is a horrible waste of taxpayer and tuition money) and the precariat post-docs unemployed. Sabitini is an absolute dope not to understand this.
In the school I’m at, two faculty got fired for fraud going on in the lab, over at least a decade. They were paid 1/2 million a year, and brought in 10’s of millions of grant money, for publications (20 or more) that are now thought to be largely wrong. This is incredibly wasteful use of money, that would have better spent supporting solid reproducible science.
My experience is that I need reproducible work from other papers to give me idea to launch me into new areas of investigation. If Im spending all of this time trying to repeat fradulent experiments, this is a huge waste of my time and makes me more likely to be a failed scientist, when I dont get publishable reults because I was tryi9ng to reproduce false results.
I think if this continues, all that will be left is “highly productive “labs that are producing junk, because all honest research has been driven out. That will keep the Chinese paper mills busy, which would be an outstanding employment opportunity for Mike. Mike, are you there? Care to comment?
LikeLiked by 2 people
https://royalsocietypublishing.org/doi/full/10.1098/rsos.160384
The natural selection of bad science
Paul E. Smaldino and Richard McElreath
“successful labs produce more ‘progeny,’ such that their methods are more often copied and their students are more likely to start labs of their own. Selection for high output leads to poorer methods and increasingly high false discovery rates.”
The cheery bit:
“We additionally show that replication slows but does not stop the process of methodological deterioration.”
“Improving the quality of research requires change at the institutional level.”
How is that going to happen with money-grubbing universities?
Which head of a university is going to advocate a reduction in their pay?
People at the head of universities will have partly been selected on their “productiveness”.
LikeLiked by 1 person
I recently accepted a 25% cut in pay to extend my time before I become a failed scientist to see to it that recent work I have done gets published, so I’m making now as much as the average secretary. I have no problem being self-sacrificing, but ask the average faculty member or administrator? Yea, right.
Maybe I can put on a permanent respirator, move to China, and create novel research. Like really novel stuff, that no one can reproduce, in a paper mill. Mike and I can collaborate. Will be billionaires. What fun!
LikeLiked by 1 person
“before I become a failed scientist” Huh?! You think you are not a failed scientist yet?! Poor old under paid post-doc 😦
I didn’t see that you blame non-westerners for your failures in your entire life though here, do you wanna put some brilliant comments on that topic that we never saw before? I missed your comments a lot man… I suggest instead of publishing nonsense articles that nobody will ever read, publish your whole series of comments as a really small book that everybody could handle with himself/herself like bible to read your comments and laugh a bit everyday. I bet that would attract lots of readers. Take a 100% cut in pay and extend your time to write this funny book man…
LikeLike
Always delighted to talk to you, Mike! I might call this “conversations with a confessed cheater…”
In answer to your question: I’ve had five advisors. All of them, except for one, seemed to be basically honest people. Maybe they didn’t have the ideas that led to papers, but they were creative people. One of them was a jerk, but he was honest and would be horrified if he had published fradulent work.
The one I left after a few years because there was a paper from his lab that everyone knew had fradulent data, and I began to suspect there was even more. And then he tried to use his grant getting power to get his wife promoted to tenure track (he succeeded, sadly), who could not read or write in English, let alone actually be creative enough to do research. And, of course, his huge ego. I guess he may have had a few qualities here and there, but most everybody in the lab knew about the fradulent work, hated him, and had no respect for his wife.
Where did my advisors hail from? The first four were from the US (WA, OK, MO, and NE, respectively). The bad one….just happened to be….from China. Purely anecdotal experience, of course, but it is what it is.
LikeLike
David Sabatini, fired for sexual harassment, and Michael Hall, Awardees in the 12th Frontiers of Knowledge Awards, Foundation BBVA (Bank Bilbao Vizcaya), Formal ceremony will take place on 2021, Sept 21th in Bilbao, Spain.
Former awardees were Charpentier, Doudna, Yamanaka, Joan Massague, and Robert Lefkowitz,
LikeLike
https://www.frontiersofknowledgeawards-fbbva.es/noticias/bilbaos-euskalduna-conference-centre-hosts-the-gala-concert-of-the-12th-and-13th-bbva-foundation-frontiers-of-knowledge-awards/
Indeed! Sabatini was awarded last year, but what with COVID-19, they are doing the award ceremony for last and this year together. What lark.
LikeLike
“12th Frontiers of Knowledge Awards, Foundation BBVA (Bank Bilbao Vizcaya), Formal ceremony will take place on 2021, Sept 21th in Bilbao, Spain.
Former awardees were Charpentier, Doudna, Yamanaka, Joan Massague, and Robert Lefkowitz,”
Joan Massague.
https://pubpeer.com/search?q=massague
Joan Massague mentee doing well.
https://pubpeer.com/search?q=stacy+blain
LikeLike
Billions of dollars of government funds are wasted on data duplication, fabrication and misconduct every year,around the world. Without Pubpeer and blogs like this, nobody will know about it or correct it. There are too many top labs who fake results to get ahead and bully anybody that questions their data. Shame on them.
LikeLiked by 1 person
The fact that the P-S473 24 hour + serum replacement band is substantially de-phosphorylated (3-4 fold) compared to the original duplicated band indicates that there is an endogenous dephosphorylating mechanism in the WT DLD that does not exist in the mut DLD that the authors did not address. This not only undermines the conclusion of this experiment; it undermines the reliability of the WT and mut cell lines used in figure 3 and 4! This, in my opinion, brings into doubt the conclusions of the entire paper.
LikeLike
My apologies. I was replying to the Kalaany Nature paper figure 3 above.
LikeLike
Reply to NMH
January 30, 2020
Where did my advisors hail from?
South Korea. 5 retractions. Image manipulation.
Italy. 1 retraction. Image manipulation.
Canada. Zero retractions.
U.S. Zero retractions.
U.S. Zero retractions,
U.K. Zero retractions.
Purely anecdotal experience, of course, but it is what it is.
LikeLike
The impossible happened: David Sabatini himself went to comment on PubPeer.


This case is interesting: Sarbassov & Sabatini JBC 2005
https://pubpeer.com/publications/E75D4B4461BFB679C4B8CFD287A615#3
Were these two S6K1 blots supposed to look so similar?
Sabatini and Sarbassov admit a mistake and post a new image:
That I am afraid is a shaky claim. The new image proves that the S6K1 total signal is so different from phospho-S6K1 that it is not possible to confuse them by mistake, or to innocently overlook the mistake after it happened.
LikeLike
Sabitini appears to have made a very qualitative appraisal there there is no change in S6K1. If he is such a big shot, surely there must be a densitometer around Whitehead for him to check this. Sheez.
Seems a little suspicious that this was in all of the prior data but not in the final review of the manuscript.
But in the end, Sarbassov will get a decent job somewhere, so its all fine…..
LikeLike
Sarbassov is back in Kazakhstan, at Nazarbayev University. A country famous for its values of democracy, ethics and justice.
I think you speak Russian, NMH? Write to the university, see what happens. Can’t be more corrupt than MD Anderson.
LikeLike
Hmmmm. …I dont speak Russian, but Im good with handling deadly amounts of radioactivity (got my degree in a Chem dept, of all places). Probably could pass through airports without notice. I can be charming at times. Maybe Putin could hire me as a hit man, if my collaboration with Mike in China fails (which seems unlikely)….
LikeLike
A clarification: I call it “benign neglect”. What this is is when the PI runs the lab, and doesn’t pay any attention to you and your project, but hopes you will produce something publishable. Usually no pressure in these situations from the PI, they just kind of wait for you to generate positive results, without helping you. I think this happens a lot to grad students in the US where positions are supported off teaching assistanships, so its not a big deal if the grad student gets nothing interesting. My US grad advisor definitely treated me with benign neglect, and it frustrated me he was helping others with their projects and not me, and it showed in my publication record.On the other hand, it taught me how to think independently. Trial by fire, as it were.
Where fraud happens is when you are working for a big shot PI and realize that a paper in Science/Nature/or Cell with your PI as corresponding author may actually get you a full time position somewhere. That’s where the grad student and post-doc commits fraud and the PI does not catch it due to his/her benign neglect. An example of that would be Carlos Lopez Otin (the benign neglector), where the grad students (the neglectees of poor ethical make up) make stuff up to pivot to an even more famous researcher (Daley). I call this the Croce affect.
LikeLike
Ever wonder wear Sabitini got the phrase “failed scientist”?
https://www.timeshighereducation.com/opinion/academics-should-stop-fixating-editorial-power-fallacy
He liked this article in his twitter feed.
To him, a “Failed Scientist” is someone who doesn’t make it to running his own lab, apparently. Funny, I see failed scientist as someone who either commits fraud to get ahead, or is ignorant of it going on in his or her lab, perhaps because they are receiving to may award to pay attention.
This guy apparently is a joke as an advisor, with all the crap that gets published on his watch.
LikeLike
“Failed Scientist” is also what such pompous academics call professional journal editors, behind their backs of course. You know, same people Sabatini is now pleading with to ignore the errors in his papers.
LikeLike
More Sarbassov and Sabatini cooperation.
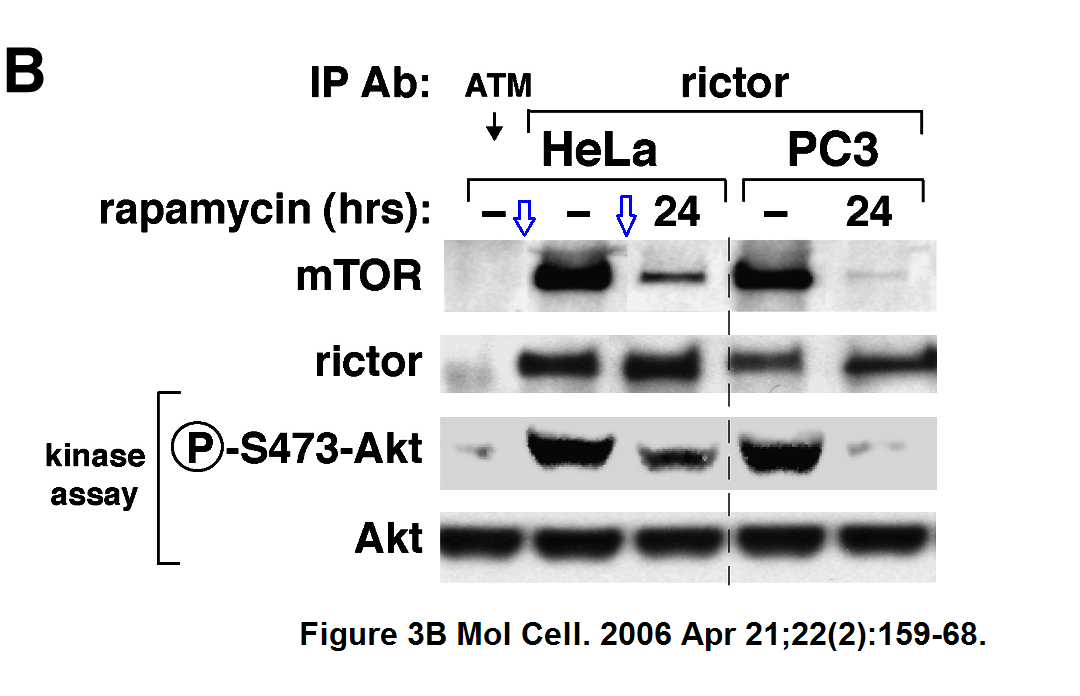
Dos D. Sarbassov , Siraj M. Ali , Shomit Sengupta , Joon-Ho Sheen , Peggy P. Hsu , Alex F. Bagley , Andrew L. Markhard , David M. Sabatini
Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB
Molecular Cell (2006) doi: 10.1016/j.molcel.2006.03.029
Note that the authors were well aware of the expectation to highlight gel splicing, see the vertical dashed line. They decided however to hide the fact that the 2nd mTOR gel band was spliced in. Why so?
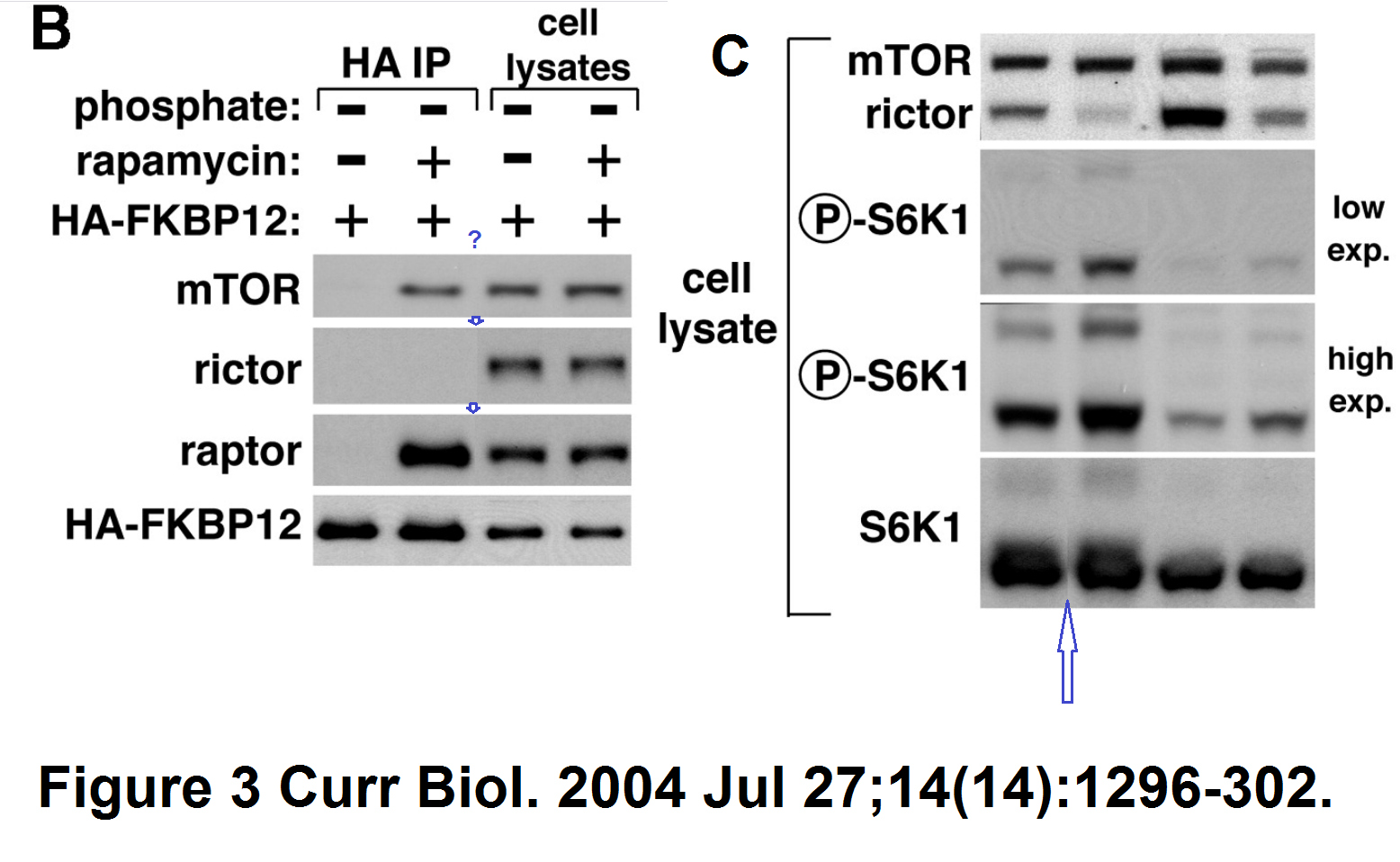
Another paper with splicing, again S6K protein making trouble:
D D Sarbassov , Siraj M Ali , Do-Hyung Kim , David A Guertin , Robert R Latek , Hediye Erdjument-Bromage , Paul Tempst , David M Sabatini
Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton
Current Biology (2004) doi: 10.1016/j.cub.2004.06.054
Here the splicing is very cack-handed, but Cell is not known as a journal to object to a bit of data rigging.
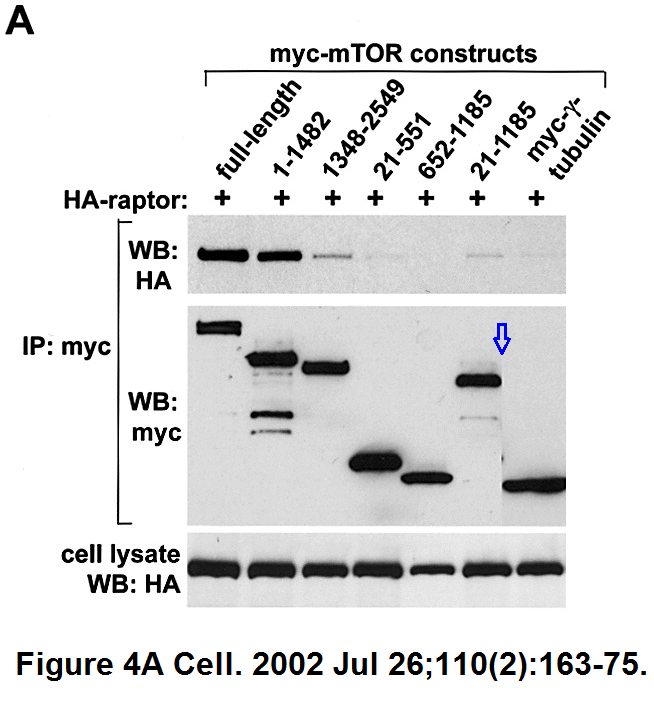
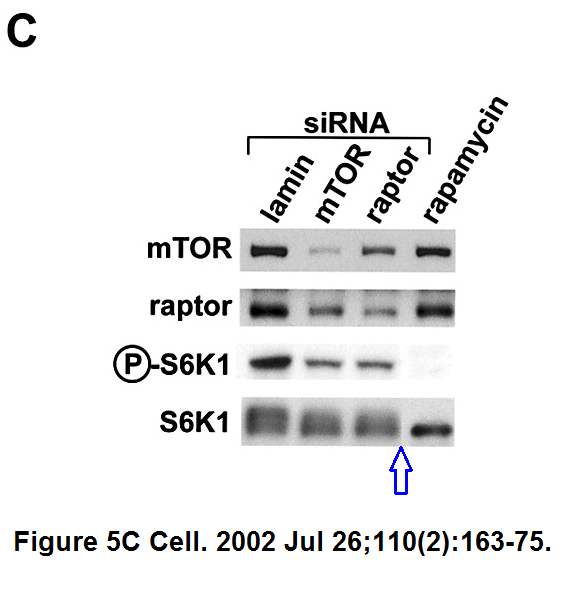
Do-Hyung Kim , Dos D. Sarbassov , Siraj M. Ali , Jessie E. King , Robert R. Latek , Hediye Erdjument-Bromage , Paul Tempst , David M. Sabatini
mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery
Cell (2002) doi: 10.1016/s0092-8674(02)00808-5
At some point it would be wise for Sabatini to distance himself from Sarbassov and to pretend to barely know that man.
LikeLike
Another weird case, a brand new Sabatini paper in Nature Medicine:
Raghu R. Chivukula, Daniel T. Montoro , Hui Min Leung , Jason Yang , Hanan E. Shamseldin , Martin S. Taylor , Gerard W. Dougherty , Maimoona A. Zariwala , Johnny Carson , M. Leigh Anne Daniels , Patrick R. Sears , Katharine E. Black , Lida P. Hariri , Ibrahim Almogarri , Evgeni M. Frenkel , Vladimir Vinarsky , Heymut Omran, Michael R. Knowles, Guillermo J. Tearney, Fowzan S. Alkuraya, David M. Sabatini
A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance
Nature Medicine (2020) doi: 10.1038/s41591-019-0730-x
This was the PubPeer comment:
First author Raghu Chivikula replied on PubPeer.
He posted a new full gel for the anti-NEK10 antibody, a longer exposure. It now makes matters worse by showing many unspecific bands previously hidden from reviewers and readers, and it turns out that what was presented as the specific NEK10 signal is just one of the weakest bands among 5. That is not trivial: NEK10 is what this paper is about according to the title, and now we finally know how specific or maybe actually unspecific that antibody tool really is. But hey, it already passed peer review.
LikeLike
Acetlylated tubulin and NEX10 are both about 55kD, while raptor is about 150. Comparing these blots it looks to me like the author is talking out of his hat here. I’m sure the antibody manufacturer won’t like this BS, since his data actually shows it’s pretty good. Sabatini seems to have difficulties recruiting competent staff, but they have sure absorbed his attitude.
LikeLike
PubPeer comment:
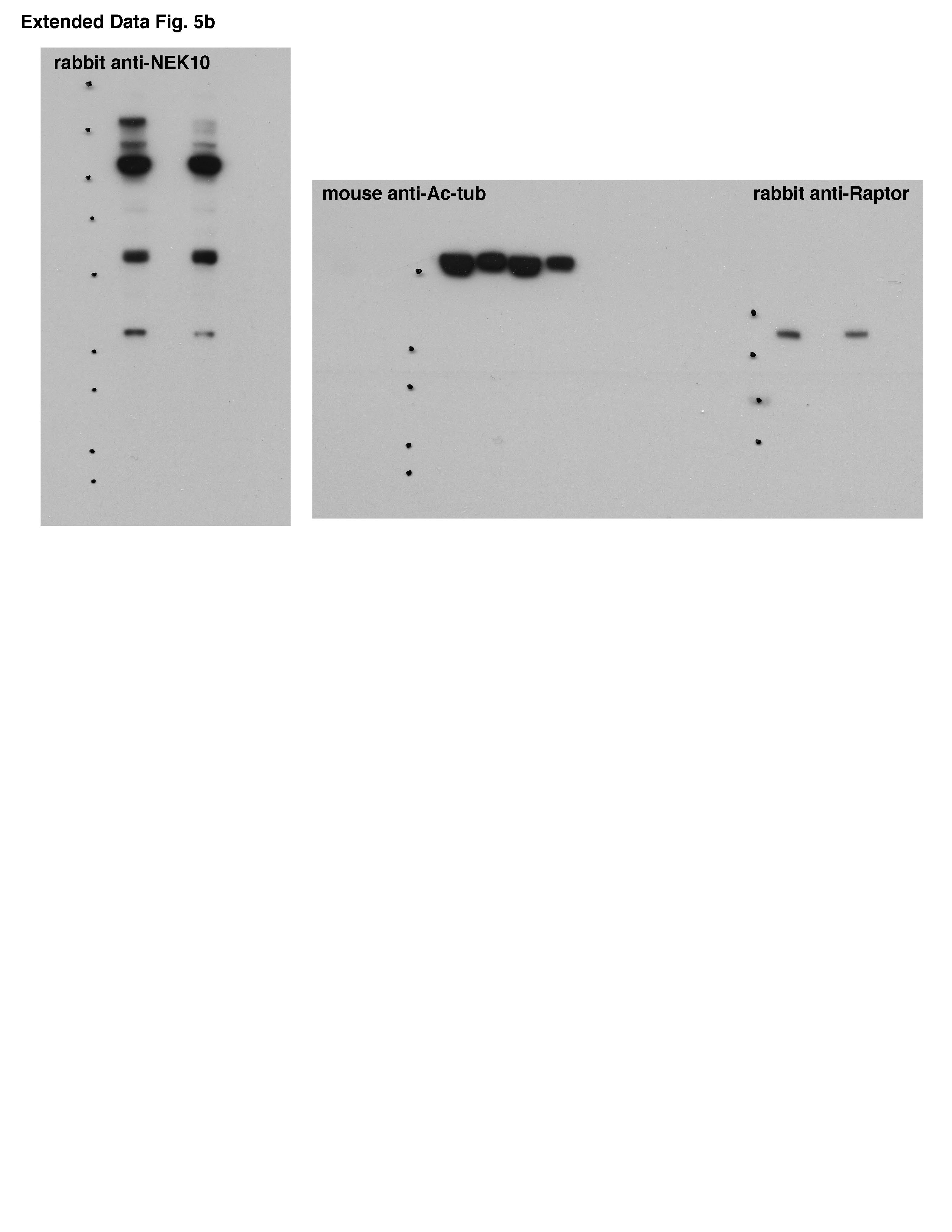
“Acetylated tubulin and NEK10 are about the same size – 55KD. Raptor is >140KD. Matching up the marker dots on the blots shown here reveals that the original poster is likely correct. The exposure shown in the original raw data is correct in showing NEK10, for which there is no discernible difference in expression between the samples shown. The author is urged to reflect on this matter.”
Good point I think. Of the 5 bands the NEK10 antibody produces the one which fits size-wise might be the real NEK10. But it doesn’t provide the desired result, there is no bloody difference!!! What to do? Let’s pick the band running up there around twice the real NEK10 size. Good enough for Nature, eh? If asked to show the gels, we refuse to label the MW marker. Brilliant! [backslaps all around].
LikeLike
I just checked on PubPeer, and the story now is that the faint band at 135kD is the “real” NEX10 – which nobody else has ever seen, including the antibody manufacturer – and the other bands are all nonspecific, including the one around 55kD that the antibody manufacturer shows on their website. Pretty impressive cherrypicking! Seems they did all sorts of roundabout tests to show that the big band is the actual one, but nobody thought to do a good old fashioned IP followed by mass spec. I still think the antibody manufacturer should go after them on this one.
LikeLike
Oh yes, indeed:
We however decided not to bother peer reviewers and readers with the details, so we showed them the 55kD band in supplementary while omitting all the other bands including the one we appointed as the only correct one. The conclusion that we made it into Nature Medicine remain unaffected.
LikeLike
This is depressing. I just prepared a paper and my advisor and he and I are always double, triple checking for errors. This should have not gotten past Sabitini. He is not doing his job, but that’s OK, hes tenured and will not suffer consequences. Since this got into Nature Medicine, the student will get a great job somewhere.
LikeLike
One more:
Yasemin Sancak , Carson C. Thoreen , Timothy R. Peterson , Robert A. Lindquist , Seong A. Kang , Eric Spooner , Steven A. Carr, David M. Sabatini
PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase
Molecular Cell (2007) doi: 10.1016/j.molcel.2007.03.003
Figure S1:
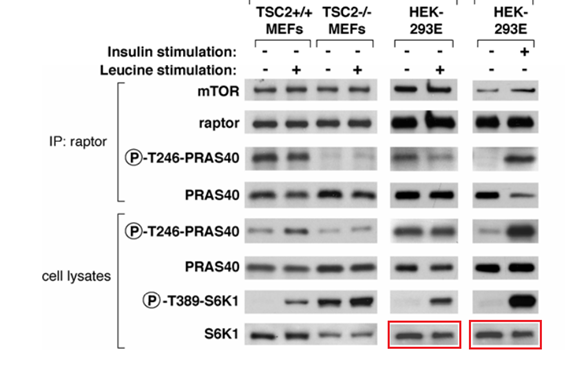
Gets worse, Figure 7B shows splicing. Sure, the authors will say it was legal in 2007. Especially since the splicing in two cases just happens to add a blank lane where no signal is supposed to be, according to the paper’s conclusions.
Yasemin Sancak is now group leader and assistant professor at University of Washington.
LikeLiked by 1 person
Pingback: Being a Great Scientist – How to (and NOT to) Handle Mistakes – Talking Tatas
More concerns about Sabatini antibody detection techniques:
Rachel L. Wolfson , Lynne Chantranupong , Gregory A. Wyant , Xin Gu , Jose M. Orozco , Kuang Shen , Kendall J. Condon , Sabrina Petri , Jibril Kedir , Sonia M. Scaria , Monther Abu-Remaileh , Wayne N. Frankel , David M. Sabatini
KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1
Nature (2017) doi: 10.1038/nature21423
PubPeer comment about a wandering protein band:
LikeLike
Wow, getting Nature papers where you don’t even know what proteins you are looking at. How Trumpian!
LikeLike
https://pubpeer.com/publications/E9FDC02781F39940EA182E3D7B1120
#8 and #9 are on another level of concern…
LikeLike
More raw data duplications.
Jason R. Cantor , Monther Abu-Remaileh , Naama Kanarek , Elizaveta Freinkman , Xin Gao , Abner Louissaint , Caroline A. Lewis , David M. Sabatini
Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase
Cell (2017) doi: 10.1016/j.cell.2017.03.023
“In Table S3.( Media profiling_24hr_R), it was odd to find that Choline and citrate presents all same values (and standard deviation) in three different conditions.”
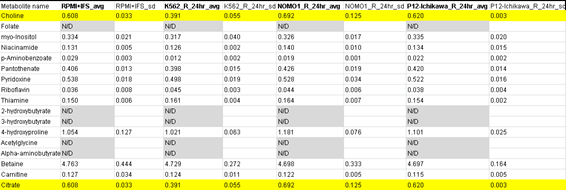

First author Jason R. Cantor, now assistant professor at University of Wisconsin-Madison, provided replacement data and explained:
LikeLike
One more!
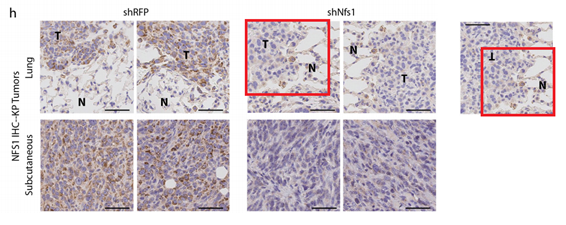
Samantha W. Alvarez , Vladislav O. Sviderskiy , Erdem M. Terzi , Thales Papagiannakopoulos , Andre L. Moreira , Sylvia Adams , David M. Sabatini, Kıvanç Birsoy, Richard Possemato
NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis
Nature (2017) doi: 10.1038/nature24637
The paper was first flagged for image duplication:
Last author Richard Possemato of New York University commented:
“when choosing representative images we inadvertently chose images of the same tumor and did not recognize the mistake because of the rotation of the image.”
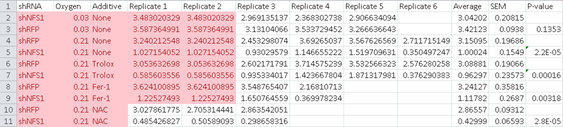
Now new concerns:
“Raw data for Figure 3B contains a set of duplicates values between replicate 1 and 2.”
LikeLike
Dear Dr. Schneider,
I’m writing on behalf of MIT’s Vice President for Research, Professor Maria Zuber, and the Senior Director for Research Administration and Compliance, Colleen Leslie, to acknowledge receipt of your email to Professor Zuber on January 31, 2020, in which you allege possible misconduct in research conducted by a member of the MIT community. In support of your concern you provide a link to a list of 44 of the subject individual’s publications in the PubPeer database, including PubPeer commentary, as well as a link to an article you authored and published on your website about the concerns.
As the Vice President for Research (VPR) at MIT, Professor Zuber initiates and oversees the review of allegations of misconduct in research conducted at MIT pursuant to MIT Policies & Procedures 10.1 (the “MIT policy”). Under both the MIT policy and federal regulations (when applicable), MIT’s receipt and review of allegations of possible research misconduct are highly confidential to protect the integrity of the review and the privacy of all persons who participate in the process, so please understand that it is very important that you not disclose to anyone, including the subject individual, that you have submitted concerns to MIT for review or that MIT is conducting a review.
If MIT needs additional information from you in order to conduct its review of the concerns you raised, we will of course contact you for assistance in the context of the review process. We will be able to update you generally about whether the review is still underway or nearing completion. Once the review is complete, you will be informed of the outcome.
MIT takes all concerns of possible research misconduct very seriously, so we thank you for bringing these concerns to the Institute’s attention.
Sincerely,
Kerry A Sousa
Compliance Officer
Office of the Vice President for Research
Massachusetts Institute of Technology
77 Massachusetts Avenue
Room NE18-901
Cambridge, MA 02139
LikeLike
Dear MIT compliance officer:
You’ve got to be kidding.
Sincerely, Everyone
LikeLike
More Sabatini irregularities.
Timothy R. Peterson , Shomit S. Sengupta , Thurl E. Harris , Anne E. Carmack , Seong A. Kang , Eric Balderas , David A. Guertin, Katherine L. Madden , Anne E. Carpenter , Brian N. Finck David M. Sabatini
mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway
Cell (2011) doi: 10.1016/j.cell.2011.06.034
I notice that the last bar, Rapa 24, is different, which makes the duplication unusual. But the first author Timothy Peterson, now group leader at Washington University, writes:
“The mistake appears to have happened when preparing the Illustrator file as the correct data appears similar such that I overlooked it. I’ve attached a corrected Figure 4A.
Thank you for finding this mistake. David will be following up shortly with the journal to correct it.”
LikeLike
Dohoon Kim, Brian P. Fiske , Kivanc Birsoy , Elizaveta Freinkman , Kenjiro Kami , Richard L. Possemato , Yakov Chudnovsky , Michael E. Pacold , Walter W. Chen , Jason R. Cantor, Laura M. Shelton , Dan Y. Gui , Manjae Kwon , Shakti H. Ramkissoon , Keith L. Ligon , Seong Woo Kang , Matija Snuderl , Matthew G. Vander Heiden, David M. Sabatini

SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance
Nature (2015) doi: 10.1038/nature14363
“In Supplemental Table 8 (Summary – peak areas), the data regarding citrate mass isotopomers M0 and M5 are the same for some – but not for all – conditions.”
Penultimate author Matt Vander Heiden, associate director of The Koch Institute at MIT replied with a lenghty message that the numbers are correct as they are and concluded:
“please contact me or the corresponding author through email and we are happy to look further to see if reliable data for this species can be extracted from the raw data, or provide you with the raw mass spectrometry data files if you have ability to process and analyze the data yourself.
LikeLike
That’s right. You, sir, need to prove there is fraud, or there is no fraud here (even though we all see it). How Orwellian.
LikeLike
Lawrence D. Schweitzer , William C. Comb , Liron Bar-Peled, David M. Sabatini
Disruption of the Rag-Ragulator Complex by c17orf59 Inhibits mTORC1
Cell Reports (2015) doi: 10.1016/j.celrep.2015.07.052
“I observed that the HA-Metap2 bands of Figure 4A and 4B are very similar.”
“After ultra-zoom them, I got more confident that they are very similar indeed. Could the authors check it please?”

LikeLike
Lawrence D. Schweitzer replied, and posted replacement data:
No mention of any corrigenda….
LikeLike
Min Wan , Karla F. Leavens , Danish Saleh , Rachael M. Easton , David A. Guertin, Timothy R. Peterson , Klaus H. Kaestner, David M. Sabatini, Morris J. Birnbaum
Postprandial hepatic lipid metabolism requires signaling through Akt2 independent of the transcription factors FoxA2, FoxO1, and SREBP1c
Cell Metabolism (2011) doi: 10.1016/j.cmet.2011.09.001
This PubPeer criticism lets one wonder how reliable the qRT-PCR results in that paper might be. Experts will understand the seriousness of concerns, and I am sure Dr Sabatini is expert enough.
“i) The Real-time PCR primers sequences provided for the TSC1 gene seems to match only introns of this gene
http://www.ensembl.org/Mus_musculus/Gene/Sequence?db=core;g=ENSMUSG00000026812;r=2:28641228-28691167
Forward primer in orange, exons in red
Reverse primer
ii) Same pair of primers were used for Acl and Gpat?
iii) The authors chose two FORWARD primers to measure Srebpc1?
http://www.ensembl.org/Mus_musculus/Gene/Sequence?db=core;g=ENSMUSG00000020538;r=11:60199089-60222581
“Forward”
“Reverse” (?)
iv) Sestrin1 forward primer is not a specific primer for this gene. A blast of this sequence with the M. musculus transcriptome look like this
LikeLike
What is it with S6 data always?
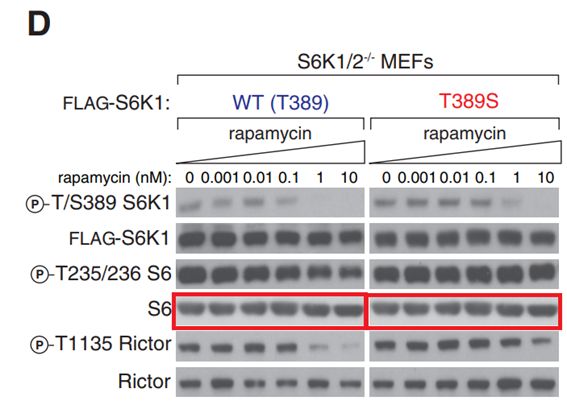
Seong A Kang , Michael E Pacold , Christopher L Cervantes , Daniel Lim , Hua Jane Lou , Kathleen Ottina , Nathanael S Gray , Benjamin E Turk, Michael B Yaffe, David M Sabatini
mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin
Science (2013) doi: 10.1126/science.1236566
LikeLike
Authors do not think a corrigendum is necessary:
“This is Seong A. Kang (currently at Navitor Pharmaceuticals, seongwoo.kang@gmail.com), first author on this paper. Thank you for your comment on Figure 3D. Upon closely examining the figure, we learned that a western blot panel for total levels of S6 was accidentally duplicated for both WT and mutant S6K1 MEFs. We have now corrected this error in the revised figure (attached) using the correct S6 blot from WT S6K1 MEFs. I’d like to emphasize that this particular change has no impact on any of our conclusions. In addition to the revised figure, we have attached the original files for the total S6 blots as well as the corresponding file information (screen capture) indicating that these files were originally created in 2012. We hope that we have adequately addressed your comment.“
LikeLike
Amateurs, really.
Scott R. Floyd , Michael E. Pacold , Qiuying Huang , Scott M. Clarke , Fred C. Lam , Ian G. Cannell , Bryan D. Bryson , Jonathan Rameseder , Michael J. Lee , Emily J. Blake , Anna Fydrych , Richard Ho , Benjamin A. Greenberger , Grace C. Chen , Amanda Maffa , Amanda M. Del Rosario , David E. Root , Anne E. Carpenter , William C. Hahn author has email , David M. Sabatini, Clark C. Chen, Forest M. White, James E. Bradner, Michael B. Yaffe
The bromodomain protein Brd4 insulates chromatin from DNA damage signalling
Nature (2013) doi: 10.1038/nature12147
“Supplemental Fig. S1c – more similar than expected
Supplemental Fig. S2c – more similar than expected”
LikeLike
Dudley W. Lamming, Gokhan Demirkan , Joan M. Boylan , Maria M. Mihaylova , Tao Peng , Jonathan Ferreira , Nicola Neretti, Arthur Salomon, David M. Sabatini, Philip A. Gruppuso
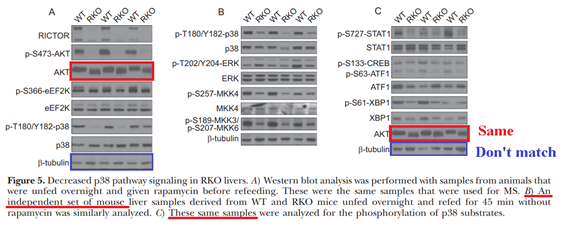
Hepatic signaling by the mechanistic target of rapamycin complex 2 (mTORC2)
The FASEB Journal (2014) doi: 10.1096/fj.13-237743
“Figure 5 – possible duplication in AKT bands in a different set of animals according the figure legends. Could authors check it please?”
LikeLike
Work for very famous advisor at “top school”, get CNS paper (in part b/c you are working for someone famous at a “top school”) => faculty position overseeing cheap labor. Sketchy data found after you get your position overseeing cheap labor => nothing happens. Yes, crime (or incompetence) does pay in science.
LikeLike
Faking pays spectacularly well in art, too. https://www.tvo.org/video/documentaries/there-are-no-fakes
LikeLike
The answer is blowing in the wind…
https://www.quora.com/How-does-David-Sabatini-MIT-manage-to-publish-so-many-high-impact-cell-biology-articles-in-a-year
LikeLike
Conclusions not affected by image fakery?
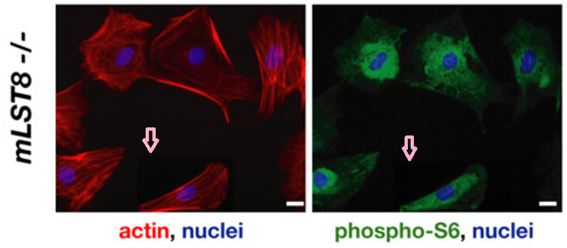
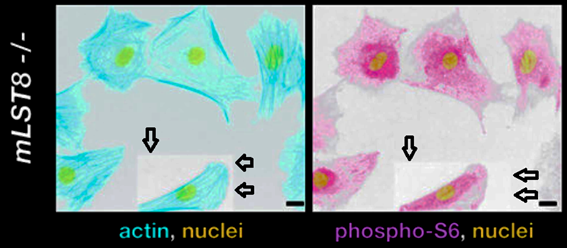
David A. Guertin, Deanna M. Stevens , Carson C. Thoreen, Aurora A. Burds , Nada Y. Kalaany, Jason Moffat , Michael Brown , Kevin J. Fitzgerald , David M. Sabatini
Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1
Developmental Cell (2006) doi: 10.1016/j.devcel.2006.10.007
LikeLike
“Michael Hall and David Sabatini will receive the Sjöberg Prize 2020”
The mTOR field is really something, hum?
Michael Hall Pubpeer CV:
https://pubpeer.com/publications/B7C09F276E1BDE265580348F41F66C
https://pubpeer.com/publications/5492B99F7D3BB99213D1FCF47784C3
https://pubpeer.com/publications/41426EFE98454CC3287C930E6668AE
https://pubpeer.com/publications/B8412948A008F8644DB2E9F536A241
https://pubpeer.com/publications/8B158F873C2D34C54E921DCB50582B
LikeLike