The Plant Cell is the most influential plant science journal, and in 2015 it stood in the centre of the biggest fraud affair in plant sciences in recent years: that of Olivier Voinnet. As PubPeer evidence started to emerge, the US scientist Vicki Vance spoke out and told a story how she was dismissed as peer reviewer by Plant Cell in late 2003, after she caught Voinnet and his right-hand man Patrice Dunoyer manipulating data. The paper was published as Dunoyer et al 2004, and when the scandal broke in 2015, and Vance published her unwelcome Plant Cell peer review and this Open Letter, it became Voinnet’s first ever retraction, seven more followed until the scientific community decided to pull a plug on the affair, and to reform Voinnet as a role model of research integrity.
Did the journal learn an important lesson from that affair? Yes, it did: carry on and don’t listen to whistleblowers. An attitude one expects from greedy Elsevier outlets like Cell, but not from a community-driven scholarly society journal (Plant Cell is published by American Society of Plant Biology, ASPB). It might be not a coincidence that Plant Cell Senior Editor and Voinnet’s colleague from CNRS IBMP in Strasbourg, Pascal Genschik, himself has a serious PubPeer record of manipulated data in his papers, one of them even in Plant Cell. Genschik recently contributed another paper to his journal, with reformed Voinnet as key co-author. Below I will show you even more examples of data fakery in Plant Cell and/or by Plant Cell editors, and because science is a village, all are somehow connected to Voinnet affair. But the real common aspect is: the journal doesn’t seem to care. All my emails went unanswered. The authors there are some very senior figures in the field, including Voinnet’s investigator Francis-Andre Wollman and his former mentor David Baulcombe, as well as Plant Cell review editors Ortun Mittlesten Scheid and even the Editor-in-Chief appointed in 2015, Sabeeha Merchant.
David Baulcombe, formerly with The Sainsbury Lab in Norwich, now in Cambridge, UK, is a giant of plant sciences, one of the discoverers of siRNA gene silencing, for which he and his PhD student Olivier Voinnet were expected to receive the Nobel Prize. It’s a pity it didn’t happen, the Voinnet fraud affair would have become even more exciting then. One version of what happened went that Baulcombe trusted Voinnet too much, another version was that Baulcombe knew of Voinnet’s devious side but arrogantly assumed to have control over him. The new official version from the responsible University of East Anglia is that Voinnet never intentionally faked any data in his PhD thesis under Baulcombe, which by extension means their papers containing same figures were retracted (or embarrassingly corrected) unjustly.
In any case, it seems it happened again in Baulcombe lab, and Voinnet had nothing to do with it.
Case 1
Padubidri V. Shivaprasad, Ho-Ming Chen, Kanu Patel, Donna M. Bond, Bruno A.C.M. Santos, David C. Baulcombe
A MicroRNA Superfamily Regulates Nucleotide Binding Site–Leucine-Rich Repeats and Other mRNAs
Plant Cell, 2012. DOI: https://doi.org/10.1105/tpc.111.095380

The first author PV Shivaprasad is now group leader in Tata Institute of Fundamental Research in Bangalore, India, and recently published another paper in Plant Cell, now from his own lab.
Also the following case comes from a very esteemed laboratory, that of Arthur Grossman at Carnegie Institution for Science in Stanford, USA.
Case 2
Wenqiang Yang, Claudia Catalanotti, Sarah D’Adamo, Tyler M. Wittkopp, Cheryl J. Ingram-Smith, Luke Mackinder, Tarryn E. Miller, Adam L. Heuberger, Graham Peers, Kerry S. Smith, Martin C. Jonikas, Arthur R. Grossman, and Matthew C. Posewitz
Alternative Acetate Production Pathways in Chlamydomonas reinhardtii during Dark Anoxia and the Dominant Role of Chloroplasts in Fermentative Acetate Production
Plant Cell. 2014 doi: 10.1105/tpc.114.129965

The same first author Wenqiang Yang, now professor at Key Laboratory of Photobiology, at Chinese Academy of Science in Beijing, is also responsible for this paper in PNAS:

Wenqiang Yang , Wittkopp TM, Li X, Warakanont J, Dubini A, Catalanotti C, Kim RG, Nowack EC, Mackinder LC, Aksoy M, Page MD, D’Adamo S, Saroussi S, Heinnickel M, Johnson X, Richaud P, Alric J, Boehm M, Jonikas MC, Benning C, Merchant SS, Posewitz MC, Grossman AR
Critical role of Chlamydomonas reinhardtii ferredoxin-5 in maintaining membrane structure and dark metabolism
PNAS (2015) doi: 10.1073/pnas.1515240112
There seems to be a common pattern. You also might recognise one of the authors as the Plant Cell Editor-in-Chief, Sabeeha Merchant. I alerted her to this problem, but received no reply. Same Carnegie authors (without Merchant) produced another paper, Wittkopp et al Plant Journal 2018, and it also contains a duplicated image of a blank field. One author there is Francis-Andre Wollman, a top scientist in France and secret investigator of both Olivier Voinnet and Catherine Jessus (with exactly opposite results vis-a-vis the same kind of data manipulation, read here).
And since we mention Wollman, this is a good occasion to remind once again of this Plant Cell paper which apparently cannot be corrected for even a minor-looking duplication. The Chicken of the Dishonour Legion Wollman cannot be associated with manipulated papers, or the French Republic might collapse or something.
Case 3
Alizée Malnoe, Fei Wang, Jacqueline Girard-Bascou, Francis-André Wollman, Catherine De Vitry
Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions
The Plant Cell (2014) doi: 10.1105/tpc.113.120113
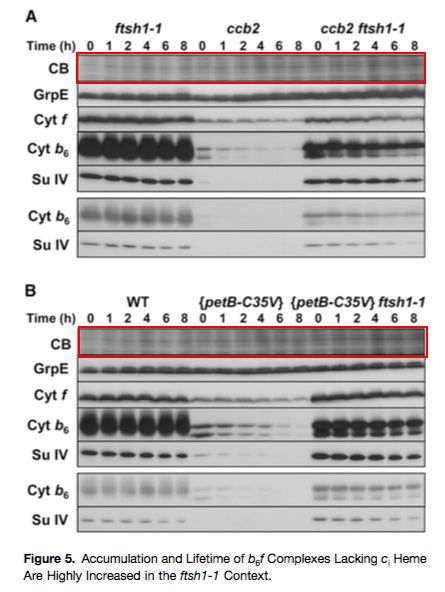
The following story also has no direct connection to Voinnet affair, only a circumstantial one.
Case 4
Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, Vaucheret H, Mittelsten Scheid O
Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 2004 DOI: https://doi.org/10.1105/tpc.018754
The penultimate author Herve Vaucheret almost became Voinnet’s PhD mentor in 1990ies, but Voinnet used that visit and its productive discussions about gene silencing as an inspiration to go to David Baulcombe’s lab instead, to work on siRNA. The corresponding last author Ortun Mittelsten Scheid is review editor at Plant Cell and group leader at the Gregor Mendel Institute (GMI) of the Austrian Academy of Sciences in Vienna. This GMI is also where a former Voinnet PhD student, Arturo Marí-Ordóñez, was recently recruited as group leader based on a Nature Genetics paper which even Voinnet himself admits was manipulated.
In March 2017, a reader of my site shared with me this evidence of some retouching activities in Figure 2A, as if someone applied a Vorwerk vacuum cleaner to the second gel lane and created a repetitive carpet pattern.
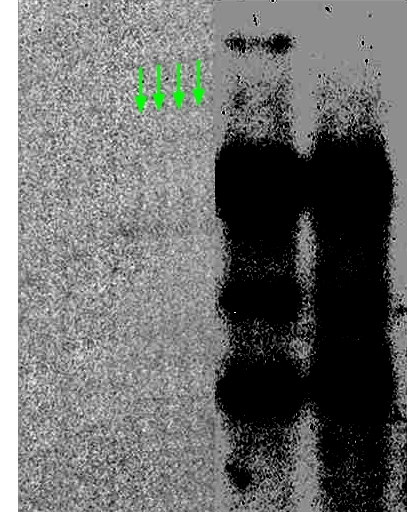
I informed Mittelsten Scheid right away, certain she will want to investigate what happened. Instead, she was apparently more interested to find out who my source was, since she merely replied:
“The reader is welcome to contact me directly”
Plant Cell Editor-in-Chief Merchant did not reply, to none of my communications in this or any other case. The next address was the Gregor Mendel Institute, and they accepted my notification on this paper and on the aforementioned Nature Genetics paper by their new group leader, a Voinnet graduate. 6 days later, on 10 September 2017, Head of Science Support Matthew Watson educated me how research integrity works:
“We have full confidence in the scientific integrity of Drs. Mittelsten Scheid and Marí-Ordóñez. If you have concerns about specific papers, please first contact the corresponding authors, then the journals, then the institutions involved (in that order). The GMI had no involvement with the papers you list.”
Funny how GMI has much involvement with these same papers when deciding whom to recruit or when applying for grants, but not when problems arise. I then notified the Austrian Agency for Research Integrity (ÖAWI), a new central authority established in Austria. They did not dismiss the case like GMI did because the Mittelsten Scheid paper came from Switzerland, but began an investigation.
On 31 January 2019 I received this “final letter” from ÖAWI:
“A forensic analysis of the originals provided by the authors at the time of submission has not confirmed this suspicion: not only in Fig. 2, but also in Fig. 6 and 8 several blots each were merged together.
The dividing lines between the blots were presumably deliberately blurred to avoid harsh contrasts. The corresponding author, Dr. Mittelsten Scheid, has confirmed this approach.
All the experts interviewed stated that such a practice was common at the time of the submission of work (2004) and not considered scientifically incorrect. Signs for further manipulation of the blots with the aim of deceive were not found”.
On 11 February 2019, the ÖAWI retracted their statement that the borders were blurred deliberately:
“This second sentence is wrong: the three illustrations show no evidence of blurring, rather are the dividing lines sharply distinctive. The author of the forensic report writes: “Although a number of images include gel lanes that have been spliced together, this was common practice at the time and I do not consider it inappropriate. More to the point, there is no evidence in this assessment for any inappropriate manipulation of any of the images in the paper.”
I then explained to ÖAWI again that the main issue here was not lane splicing, but the Vorwerk-vacuumed second gel lane. Like, what was wrong with the signal there that it had to be removed and that now everyone refuses to even look at it?
On 28 February 2019 I received this final final decision:
“Here again the summary of our opinion: Also Fig. 2A was the subject of the forensic report, no indications for irregularities were found there.
In our last letter, there was a misunderstanding: Dr Mittelsten Scheid only confirmed that in the said figures in each case several rows of different blots were put together. She has never testified or confirmed she or her fellow authors performed further manipulations (such as blurring of the dividing lines), she rejects this suspicion vehemently.The cause of the fine lines observed by your reader (marked by the green arrows) cannot be clarified, since the original material remained due to a contractual obligation in the relevant former laboratory in Basel, and the plant research groups have been terminated there since a long time. Should the same guidelines apply in Basel as in Austria, nobody would be obliged there to store original data for more than 10 years.
To conclude, I repeat that the ÖAWI Commission did not find any indication of scientific misconduct.”
This unavailability of original data apparently explains why nothing can be wrong with that Figure. Genius.
The Senior Editor and his colleagues
None of the above seems to bother the journal. Because Plant Cell is an elite outlet, the authors and editors there are some serious heavyweights whose labs apparently cannot be associated with data manipulation willy-nilly. Remember the effort University of East Anglia went through to prove Voinnet’s PhD thesis under Baulcombe’s was perfectly fine, despite containing exactly same fraudulent figures as their common retracted and corrected papers?

Which brings us to the Senior Editor at Plant Cell, Pascal Genschik. He knows how damaging research fraud is to science, his IBMP in Strasbourg became a focus of ridicule and suspicion because of Voinnet and Dunoyer. The latter was appointed the scapegoat for his own crimes as well as for the crimes of everyone else and sent away to the other side of the world, where he originally came from: to New Caledonia. That was necessary because so many senior researchers at IBMP, including the director Laurence Drouard, dabble as connoisseurs of the high art of data fabrication, as their papers and supervised PhD theses demonstrate. Recently, another IBMP group leader Wen Hui Shen was reported on PubPeer by a reader of my site for publishing shady data. He even has a common paper with Genschik questioned there. And this Plant Journal paper from 2002 is particularly entertaining because there is only one author on it, Wen Hui Shen. Nobody to scapegoat here, not even Dunoyer!
But now, would you like to see what Senior Editor Genschik published in his own journal?
Case 5
Arnaud Capron , Olivier Serralbo , Katalin Fülöp , Florian Frugier , Yves Parmentier , Aiwu Dong , Alain Lecureuil , Philippe Guerche , Eva Kondorosi , Ben Scheres , Pascal Genschik
The Arabidopsis anaphase-promoting complex or cyclosome: molecular and genetic characterization of the APC2 subunit
The Plant Cell 2003. doi: 10.1105/tpc.013847

One generally does not defecate where one eats, but Genschik did. Admittedly he only became Plant Cell editor the next year, in 2004, but still. One can at least tidy up after oneself even if the deed is already done. But here it is, this Capron et al 2003, uncorrected and it might never will be.
To be fair, the EMBO member Genschik published even worse stuff in a different journal, in PNAS: in Derrient et al 2012:

Incidentally, one of key co-authors on that paper is Plant Cell review editor Karin Schumacher, professor at University of Heidelberg. She recently published a paper in eLife, Patir-Nebioglu et al 2019, which might be the first serious case of publicly recorded data manipulation that young journal has to deal with, under its new Editor-in-Chief, the Open Access enthusiast Michael Eisen.
It’s not like Genschik didn’t care at all. He used to care: he made an effort, by ordering the responsible author of Derrient et al PNAS 2012 back to the Strasbourg lab, presumably in search of the original data, and it looked as if he intended to publish a corrigendum. But in the end, it seems, PNAS didn’t mind. They don’t mind many things, also specifically in plant sciences.
It’s not like only the Plant Cell learned nothing from Voinnet affair. Hardly anyone learned anything, it seems. Most scientists do not have the ethics of Baulcombe’s colleague Jonathan Jones of The Sainsbury Lab. In the wake of Voinnet scandal, Jones investigated and retracted two of his own papers, after discovering data manipulations by his then-postdoc Susana Rivas. One of them was in Plant Cell:
Case 6
Susana Rivas, Tina Romeis, and Jonathan D. G. Jones.
The Cf-9 Disease Resistance Protein Is Present in an ∼420-Kilodalton Heteromultimeric Membrane-Associated Complex at One Molecule per Complex.
Plant Cell 2002. doi: 10.1105/tpc.010357. Retraction: February 2017.
Four figures in that paper were found heavily manipulated, yet none of that ever was flagged on PubPeer, nobody knew. There was no pressure on Jones, from nobody but his own ethics. Now look at his colleagues and their Plant Cell papers.

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism will be most appreciated!
€5.00


Another amazing issue in France right now is the obsession with “Shanghai ranking” that leads many authors to affiliate to up to 8 institutions at the same time:
Example (stumbled by chance) from this group:
https://www.sciencedirect.com/science/article/pii/S0168945215300625?via%3Dihub
where the institutions are:
Institute of Plant Sciences Paris-Saclay (IPS2), CNRS, INRA, Université Paris-Sud, Université d’Evry, Université Paris-Diderot, Sorbonne Paris-Cité, Université Paris-Saclay.
There are much more examples like this from French authors where a mega affiliation is used (check PubMed).
How one can be affiliated to up to 5 different universities (Université Paris-Sud, Université d’Evry, Université Paris-Diderot, Sorbonne Paris-Cité, Université Paris-Saclay) and 3 different research centers (IPS2, CNRS, INRA) at the same time?
Ridiculous, isn’t it?
Some amateurish Chinese guys succeeded to make an institutions’ game in France where authors are asked to mention up to 10 institutions in their papers to boost their ranking positions in the so-called Shanghai ranking!
LikeLike
To start, I completely agree about how ridiculous that Shanghai ranking is. As for the affiliations, those are the result of some particularities of the French academic system. I myself used to have 3 affiliations (CNRS, U-Bordeaux 1, and U-Bordeaux 2) until the Bordeaux universities merged, so now I have only 2 (CNRS and U-Bordeaux). My employer is the CNRS. The institute in which I work is at the University of Bordeaux. Our Institute is a joint “unité” between CNRS and U-Bordeaux. Because of Shanghai, both insist that if I publish, I give two affiliations. If I publish together with a colleague from, say, INSERm, who is located at another university, the two of us already have 4 affiliations. If the colleague is in an institute supported by 3 institutions, we’d have 5. I have CNRS colleagues who work in an INSERM lab and the other way around. They have to list both CNRS and INSERM as affiliation… It’s not that much different from somebody who is a Howard Hughes investigator in the US, that person will list both the Howard Hughes Institute and the institution where he is located as affiliations. It’s just a peculiarity of the French system that you can have 3 or even more such affiliations. I can even see the logic behind me having 2 affiliations, as my work is possible because I earn a salary (CNRS) and have a place to work (U-Bordeaux) and on top of that get some funding (not much, but still) towards my research, for which I don’t have to write cumbersome grant applications. Hope this didn’t get too confusing…
LikeLike
This is my own opinion about the case 1 (Shivaprasad et al., 2012, Plant Cell).
1- There clearly is an undeclared splicing, which already is a problem.
2- but more importantly, the spliced square is a “copy/paste” from an empty region from the very same image.
After looking at the conflicting image and the original scan recently provided by the first author in pubpeer I can imagine what is going on here:
the membrane was first probed against tomato miRNA472
then membrane was stripped to erase the former signal from the tomato miRNA472
then the (clean??!!) membrane was reprobed against A. thaliana miRNA472. And here is the problem: the former signals from tomato miRNA472 in the first two lanes (two tomato ecotypes) did not disappear when he did the stripping!! Such a pity and such a problem if you do not want to repeat the whole process.
To avoid the repetition of blotting, it seems to me that they decided to copy/paste an empty region from the image to hide the original signals from tomato miRNA472 in these two lanes.
Is it a good practice? NOT, IT IS NOT.
By the way, in the original image provided by the first author in pubpeer he wrongly label “Rice” where it should say “A. thaliana”, and viceversa.
LikeLike
I guess the reader referred to an author reply here: https://pubpeer.com/publications/1831E5D553C9D34D9384C8F8407BF2#2
LikeLike
Now this is unfortunate.
Padubidri V. Shivaprasad , Thomas Hohn, Rashid Akbergenov

Biochemical Requirements for Two Dicer-Like Activities from Wheat Germ
PLOS One 2015
https://doi.org/10.1371/journal.pone.0116736
Conceived and designed the experiments: PVS TH RA. Performed the experiments: PVS RA. Analyzed the data: PVS RA. Contributed reagents/materials/analysis tools: PVS RA. Wrote the paper: PVS TH RA.
Figure3B
LikeLike
The author has now admitted there is a “discrepancy” with the blot. Contacting the journal to see what can be done about it. May we suggest a retraction? I would try that with PubPeer, but I’m tired of their comment sifting.
LikeLike
PubPeer moderation is indeed peculiar. One could take issue with this author reply, by Oscar Portillo Moreno:
“Pinche estúpido Dedica a trabar y ys deja de estar chingando Imbécil Estudia y luego te explico”
https://pubpeer.com/publications/8D16E32378DE46D37140B5FC5EA575#2
Only that PubPeer don’t seem to take issue…
LikeLike
The miR At 472 blot is a weaker exposure and/or post stripped and/or photoshopped version of the miR SI 472 blot. All bands have directly proportional intensities, as do the black spots on the left side. If you take the SI blot and drop the intensity, then increase contrast and put through jpg saving a few times you can get that same strange mottling and pixellation. I think he’s having you on, old son. BTW, the author is at the NCBS in Bangalore. It’s under the Tata umbrella, but an independent outfit. Much higher standards than where he did his training from my experience.
LikeLike
Almost 2 years after it was advertised on Pubpeer, this other “The Plant Cell” paper didn’t get any update
https://pubpeer.com/publications/C0EC30834400B78277B22483392C96#fb121780
LikeLike
Over 2 years after these two “The Plant Cell” papers were advertised on Pubpeer, still no updates.
https://pubpeer.com/publications/388CA67CD6E3649D00DA8DC0C5EE79
https://pubpeer.com/publications/8FBEFC14BF3ED158BE0CB2A923801A
LikeLike
Many more in TPC like this one (only one example):
https://pubpeer.com/publications/67195110882D5F56D73DAC906EB2EE
LikeLike
And another one. A minor issue, very likely an oversight. But how can I alert the corresponding author and journal’s EiC Sabeeha Merchant if she is refuses all communication?
Eugen I. Urzica, David Casero, Hiroaki Yamasaki, Scott I. Hsieh, Lital N. Adler, Steven J. Karpowicz, Crysten E. Blaby-Haas, Steven G. Clarke, Joseph A. Loo, Matteo Pellegrini, Sabeeha S. Merchant

Systems and Trans-System Level Analysis Identifies Conserved Iron Deficiency Responses in the Plant Lineage
The Plant Cell 2012. DOI: https://doi.org/10.1105/tpc.112.102491
LikeLike
Well congrats Leonid, it seems your article triggered a correction for this article:
http://www.plantcell.org/content/early/2019/04/05/tpc.19.00217
LikeLike
Pingback: Sally Assmann, right EiC choice for Plant Cell? – For Better Science
A reader alerted me to this interesting case, on a 2018 PNAS paper from University of Ghent, coauthored by Pascal Genschik.

https://www.pnas.org/content/115/17/E4130/tab-article-info
In the Figure S6, authors placed labels over some peculiar bits of the gel image:
LikeLike
Case 5, aka Capron et al. 2012, just got a correction.
http://www.plantcell.org/content/early/2019/04/08/tpc.19.00223
Leonid, another nice achievement !
Kudos to the massive editorial effort, highlighted here [backets are mine]
Editor’s note: the corrected figure and accompanying text [by a senior editor] were reviewed by members of The Plant Cell editorial board.
LikeLike
Thanks guy for your time and efforts to clean up the scientific record!!
LikeLike
Pingback: Get a life with Gary Stacey – For Better Science
The above mentioned plant scientist Karin Schumacher has developed a significant PubPeer record, with 8 entries in total.
Patir-Nebioglu_et_al_2019 got retracted in March 2019, just three weeks after publication, and for which Dr. Schumacher is the corresponding author.
In the other 7 cases she is co-author.
Here are the 8 articles:
https://pubpeer.com/publications/597F4A6EC25F0EE8787C936C4E7DCD (Retracted)
https://pubpeer.com/publications/DFCD2F1DAF41F0030B9969640BD245 (Corrected)
https://pubpeer.com/publications/5FFB36C5EDC851BA90F276F889404E
https://pubpeer.com/publications/11EB737C87B9FDBA08A7532C7B0249
https://pubpeer.com/publications/F8D4ACEF7E8B4FE94E34B2124F1939
https://pubpeer.com/publications/9C1ACFC8A80FCEBC9C9B2A7A0F0E6D
https://pubpeer.com/publications/EBB2EBBEB27C9A54778A7C19CE7872
https://pubpeer.com/publications/F31552D1FB342D6BE6F5CCB73C7B40
Ironically, few months after the retraction of Patir-Nebioglu_et_al_2019, in fall 2019 Prof. Schumacher has been elected Pro-Rector for Quality Development at Heidelberg University.
LikeLike
Pingback: Research Integrity Fail in Austria – For Better Science
Pingback: Rick Vierstra probably meant well – For Better Science