Sonia Melo, the Portuguese cheater scientist and her former US-boss Raghu Kalluri issued some days ago a biorxive preprint, which sole purpose is to defend their discredited Nature paper from 2015. There, they originally claimed to have found a unique biomarker for early pancreatic cancer, a much hailed promise to save lives of many cancer patients. However, soon it was found out that the results were not reproducible, the Nature paper Melo et al, 2015 contained evidence of data manipulation (just like other Melo publications with Kalluri and her PhD boss Manel Esteller). The antibody, on which the central evidence for the allegedly unique pancreatic cancer biomarker glypican 1 (GPC1) was based, proved to be delivering staining artefacts; the vendor Thermo Scientific soon discontinued it. Even Kalluri seemingly distanced himself from his results.
Now in their new preprint, Melo and Kalluri claimed to have perfectly reproduced their original 2015 GPC1 results with a new antibody, which however proved to be likely exactly the same as the old one, but sold by a different vendor. As soon as this became known, the authors issued a new preprint version just two days later, featuring yet another entirely new GPC1 antibody, with an utterly new set of results to complement the 6 day older ones. However, also these results are most likely useless. The authors namely freely admit in their preprint method description to have intentionally manipulated their flow cytometry (FACS) data to obtain a positive signal specifically where needed. It seems that one full professor and three research group leaders have absolutely no understanding about data integrity in flow cytometry (or maybe even in research in general). Obviously, they simply adjust the FACS settings for each sample in an analytic row any way it pleases them until they see a result they like. As they don’t even hide it, they seem to think this is the proper way to do science.
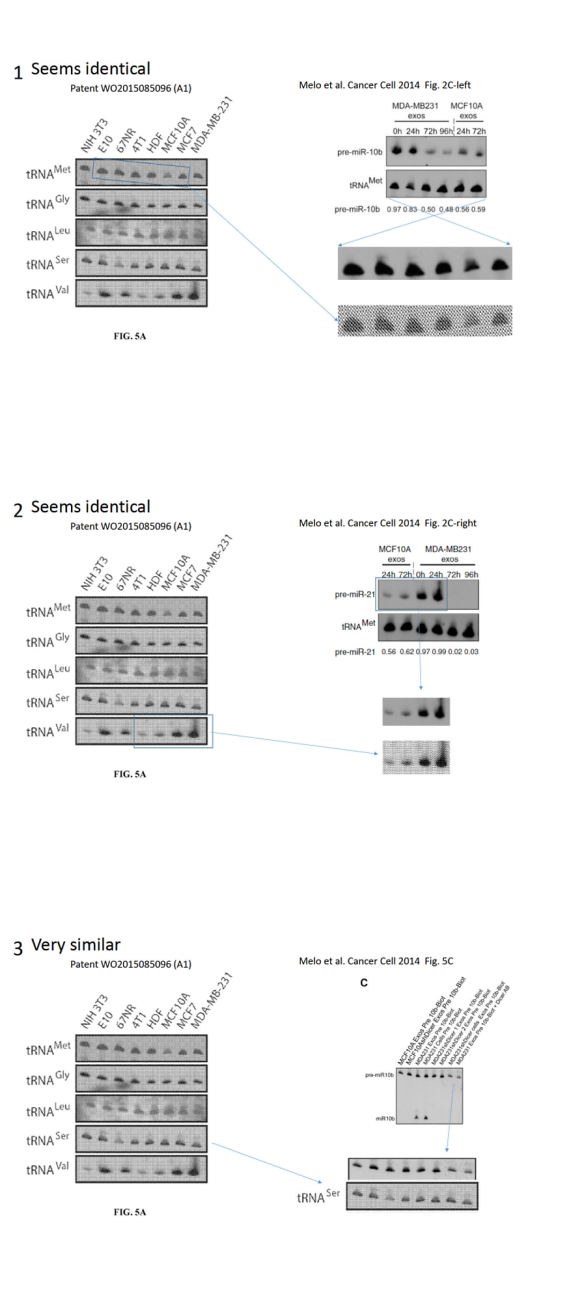
 Melo is a European scandal scientist, after she had to retract a paper (Melo et al, Nature Genetics, 2009), and lost her EMBO Young Investigator funding last year, for data manipulations (see my report here). Nevertheless, her Portuguese employer Instituto de Investigação e Inovação em Saúde (I3S) in Porto re-installed her as group leader after an investigation which can only be described as a white-washing travesty (see my report here). Kalluri is Melo’s mentor from her postdoctoral period at MD Anderson at Texas. Their common publications were flagged on PubPeer for clear evidence of data manipulations, most recently data from a Cancer Cell paper Melo et al 2014 was found to be apparently re-used in a Melo & Kalluri patent, in a different context. Kalluri himself was never investigated by his employer MD Anderson, which is easy to explain with the fact that the Texan cancer research centre is a stakeholder in his company, Codiak Biosciences, which raised in this way at least $80 Million investment (see my report here). And the main product Kalluri and MD Anderson intend to market with Codiak is… Melo’s findings of GPC1 as pancreatic cancer biomarker, published in Melo et al, Nature 2015.
Melo is a European scandal scientist, after she had to retract a paper (Melo et al, Nature Genetics, 2009), and lost her EMBO Young Investigator funding last year, for data manipulations (see my report here). Nevertheless, her Portuguese employer Instituto de Investigação e Inovação em Saúde (I3S) in Porto re-installed her as group leader after an investigation which can only be described as a white-washing travesty (see my report here). Kalluri is Melo’s mentor from her postdoctoral period at MD Anderson at Texas. Their common publications were flagged on PubPeer for clear evidence of data manipulations, most recently data from a Cancer Cell paper Melo et al 2014 was found to be apparently re-used in a Melo & Kalluri patent, in a different context. Kalluri himself was never investigated by his employer MD Anderson, which is easy to explain with the fact that the Texan cancer research centre is a stakeholder in his company, Codiak Biosciences, which raised in this way at least $80 Million investment (see my report here). And the main product Kalluri and MD Anderson intend to market with Codiak is… Melo’s findings of GPC1 as pancreatic cancer biomarker, published in Melo et al, Nature 2015.
There is therefore much at stake, and Kalluri’s standing at MD Anderson is likely not as safe as it used to be. The director of this huge Texan cancer research institute, Ronald DePinho, who originally recruited Kalluri as a star asset from Harvard, resigned from his position in March 2017, due to “financial problems, a large layoff, and a scathing audit that raised questions about his spending practices and management”. Is MD Anderson’s multimillion investment into Kalluri’s Codiak one of these financial problems? One could also speculate that DePinho’s own PubPeer record of suspicious data did not go entirely unnoticed in this context.
My site featured a third-party post-publication peer review of that Melo et al, 2015 paper, which took apart its science and revealed it as artefacts at best (see article here). The Nature publication of the zombie scientist Melo became a zombie paper, hardly anyone takes its claim of GPC1 as unique pancreatic cancer biomarker seriously anymore. Even Kalluri himself commented in the context of a paper by a different group (which analysed a set of 5 proteins):
“What’s exciting about the study is that it further favors the belief that one biomarker by itself may not be able to successfully identify a disease”.
Now however, Kalluri decided to fight the last battle for his Nature paper and the Codiak investments. The preprint is titled:
Multiple antibodies identify Glypican-1 on serum exosomes from patients with pancreatic cancer
Its authors are:
Sonia Melo, the zombie scientist of I3S in Porto; Christoph Kahlert, clinician at the University Clinic Dresden, Germany, Valerie LeBleu, assistant professor at MD Anderson, and Raghu Kalluri, Chair of the Department of Cancer Biology at the same Texan cancer research centre. All four have a common interest in studying exosomes (extracellular lipid membrane vesicles) and the marker proteins on them, for the purpose of cancer diagnostics. Only that they all seem to struggle with the basic principles of good scientific practice and ethics, while defending their vision of GPC1 being the unique pancreatic cancer biomarker.
While evidence suggests the samples in the Nature 2015 paper were up to 30 years old, the new Melo et al preprint suggests that US patients and healthy donors were invited to bleed for the sole purpose of saving that Nature paper:
“Serum samples were obtained from patients with pancreatic cancer. Serum samples were also obtained from patients with a benign pancreas disease and from healthy donors, who had no evidence of acute or chronic or malignant disease and had no surgery within the past 12 months. The cases were obtained under an IRB-exempt protocol of the MD Anderson Cancer Center (IRB no. PA14-0154).
Currently, there are two versions of the manuscript. The first one, from June 2nd 2017, contained this statement:
“Here we report on the use of two anti-glypican 1 antibodies for the specific detection of GPC1+ exosomes in the circulation of pancreas cancer patients (n=10) compared to healthy donors (n=9) and benign pancreatic diseases (BPD, n=2) using flow cytometry as a readout. The ThermoFisher antibody used in our initial report1 (PA5-28055) is commercially available. We also utilized antibody SAB2700282 from Sigma. Both companies described the immunogen as a recombinant fragment of human GPC1 corresponding to a region within amino acids 200 and 558. We report on the highly reproducible and strong correlation between samples evaluated with each of these antibodies”.
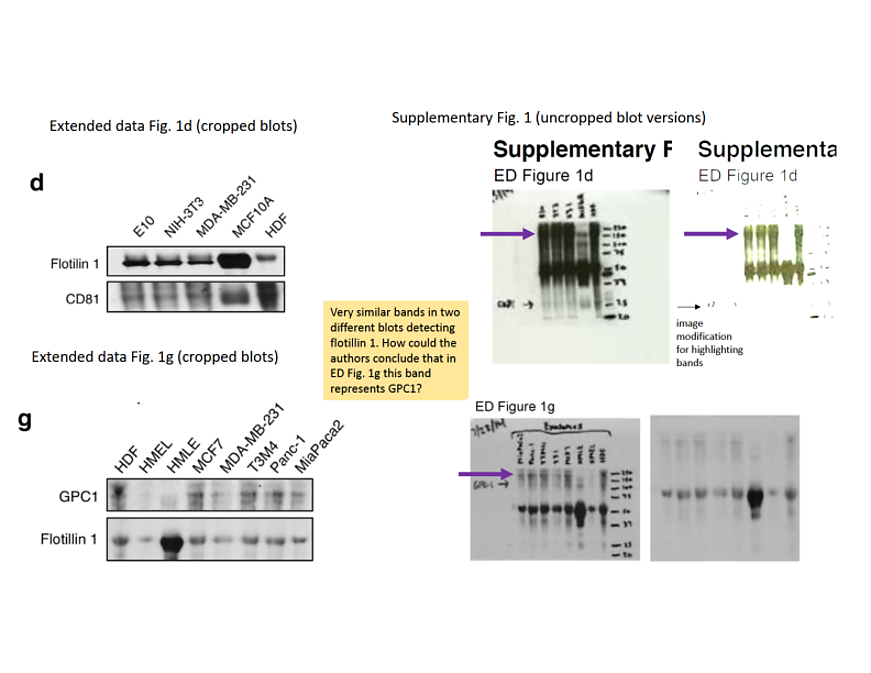
Well, in reality the Thermo Fisher product is discontinued and is not commercially available. At least not from Thermo Fisher, that is. The new Sigma Aldrich antibody namely is most likely nothing else but the old Thermo Fisher antibody (data sheet backup here), possibly after the latter vendor sold the remaining stocks of that somewhat discredited product to the former. This is clearly evidenced by the identical images of western blot, immunofluorescence and immunohistochemistry. Sigma Aldrich and its new owner Merck Millipore chose not to explain the documentation overlap to me. We have to assume however, the evidence for the specificity of the criticised Thermo Fisher antibody came from this same antibody, relabelled. But this is not the best bit.
On June 6th, I emailed Kahlert, informing him that he and his co-authors apparently used same antibody twice in order to validate one of them. I received no reply, but on June 8th, Kahlert and his 3 partners issued a new version of their preprint, which stated :
“Here we report on the use of three anti-Glypican 1 antibodies for the specific detection of GPC1+ exosomes in the circulation of pancreas cancer patients (n=10) compared to healthy donors (n=9) and benign pancreatic diseases (BPD, n=2) using flow cytometry as a readout. The ThermoFisher antibody used in our initial report1 (PA5- 28055) is commercially available. We also utilized antibody SAB2700282 from Sigma, and MAB8351 from Abnova. Both ThermoFisher and Sigma described the immunogen as a recombinant fragment of human GPC1 corresponding to a region within amino acids 200 and 558. Abnova describes the immunogen as recombinant protein corresponding to full length human GPC1”.
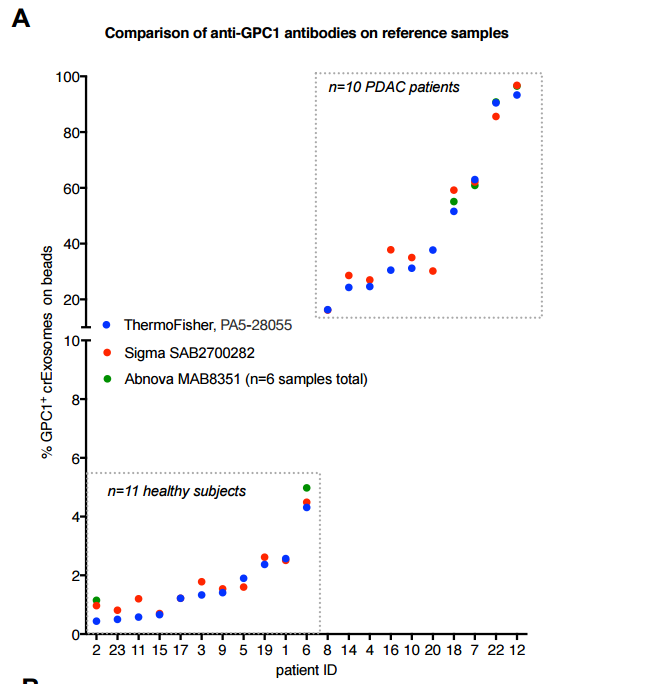
Go try making sense of that. Suddenly, an utterly new antibody (from Abnova) came from nowhere, and now it is a monoclonal antibody, which is much more target-specific than polyclonal antibodies. In two days you can hardly get an ordered reagent delivered, never mind running all analyses with it. A PubPeer critic commented like this:
“The data presented for this Abnova monoclonal seem very partial, because only concerning 6 samples of the 21 sample panel. If researchers want to check whether a staining pattern could be due to non-specific interactions, would it be the logical choice to check the entire 21 sample panel with other polyclonal antibodies, and only 6 of those samples with a monoclonal?
Antibodies have different affinities, and the number of specific plus non-specific interactions with a complex target are expected to differ substantially between a polyclonal and a monoclonal. Nevertheless, for the 6 reported samples Melo et al. seem to find virtually identical staining identities in their system (which is based on differences in staining intensity within a single peak, hence very sensitive to antibody variation). This needs some technical explanation, which is almost absent in this bioXriv paper, also because the system must have been calibrated for different secondary antibodies.
Maybe the authors should make a third manuscript version with more explanation about the use of the Abnova monoclonal”.
Indeed, how can different antibodies give identical signals? All three antibodies are plotted by the preprint authors in one single diagram. Which is only appropriate if those measurements were performed on same FACS device in parallel, under exactly same settings. Which they were not. The Abnova antibody data was added later on, when Melo et al apparently bent the time-space-continuum to create new experiments as response to my email to Kahlert. As for same flow cytometry or FACS settings, well, Melo et al provide a detailed protocol on GPC1 exosome detection, with point 23 in table 4 teaching:
“At acquisition during FC [flow cytometry, -LS], adjust the FITC voltage to ensure NC [negative control, -LS] peak (autofluorescence detected in exosomes + beads + secondary ab only) is less than 10^3 FITC log scale and tail is ~1% into the positive signal (gate set at 10^3)”.
FITC is a fluorophore which labels the secondary antibody and which signal is detected by the flow cytometry. Voltage of the FACS laser correlates to the intensity of this signal’s output. The higher the voltage, the more amplified the signal becomes; weak or unspecific staining or even the background auto-fluorescence (which every sample has) can be thus easily turned into a specific-looking signal. This is why one always needs negative controls in flow cytometry, like unstained beads or samples treated only with the fluorophore-coupled secondary antibody, to set up the device and the detection thresholds. FACS measurements of specifically-stained samples are then performed as relative to those negative controls. Every PhD student training on FACS knows not to change settings between samples which signals are supposed to be compared against each other.
But look now what the 4 elite researchers did, and they are even not ashamed to proudly announce this: they used a different voltage setting for each of their samples, between 245 mV and 440 mV. Not even two are same. Thus, the voltage for every single FACS sample was “adjusted” to get the result of GPC1 positive exosomes which the authors expected from it. A more appropriate description for this would be: data manipulation, or if done with full knowledge of consequences, research misconduct. Given that the primary antibodies were used at concentration of almost 1:10 and secondary antibody at 1:20, without such rigging the measurements for all samples (healthy and cancer patients, but also negative controls) were probably all over the place and had no scientific value or message whatsoever.
It is very logical to expect that exactly the same “adjustments” or data manipulations were applied to Melo & Kalluri’s Nature paper, where GPC1 flow cytometry was used to identify blood sera from cancer patients in such allegedly reliable manner. The peer reviewers were obviously left in the dark about it, otherwise they would have rejected the manuscript outright. An ethical thing for Nature would be to request the original FACS files and analyse their metadata sample by sample for changes in voltage settings. The next logical step would be for Nature to issue a retraction if such manipulations are indeed found, or if the raw FACS data should suddenly become unavailable. Until then, let us wait which surprises the third version of the Melo et al preprint will bring.
Update 30.06.2017. I notified the Ombudsman of the TU Dresden, Achim Mehlhorn, about the inappropriate FACS data acquisition, indicating suspected research misconduct by Christoph Kahlert (obviously there was no point to contact the host institutions of Melo or Kalluri with these concerns). The chemistry professor and former rector of the Dresden University announced to investigate, and asked a “trusted person” from the medical faculty to discuss the concerns with Kalhert. It all ended with Kahlert being acquitted from all suspicions of data manipulations, while advised not co collaborate with Melo and Kalluri again, so that his “scientific reputation” suffers no damage. Kahlert’s explanation on the need to manipulate FACS settings was referred to me by Mehlhorn as such:
“The adjustment of settings on the FACS device was explained by Dr. Kahlert that the sample heterogeneity was very large (human samples). Therefore, an individual adjustment should be made with and without antibodies so that each sample has its own control”.
Based on this discussion with Kahlert, the nameless “trusted person” then decided that there was no data manipulation and no research misconduct. I then pointed out to Mehlhorn my own well-documented experience of many years with this methodology and opined that Kahlert’s explanation made no sense and showed a lack of understanding at best. The Ombudsman Mehlhorn then attested that it was I who acted against good scientific practice. He also demanded that I keep his decision to acquit Kahlert and the entire procedure secret, despite the fact that I introduced myself as journalist from the very beginning.
I personally think that this secrecy and the academic gagging of the press is the entire problem with German academia, which also explains why we hardly ever have any misconduct here.

Donate!
If you are interested to support my work, you can leave here a small tip of $5. Or several of small tips, just increase the amount as you like (2x=€10; 5x=€25). Your generous patronage of my journalism will be most appreciated!
€5.00

Here another pancreatic cancer scandal in PubPeer who seems to generate Prizes:
https://pubpeer.com/publications/25985394
LikeLike
Interestingly the paper mentioned above comes from a lab well known as being a Dr. Kalluri’s main competitor, both of them were at Champalimaud Foundation
LikeLike
Indeed Dr. Kalluri was at Harvard (http://news.harvard.edu/gazette/story/2012/01/tumor-cells-can-prevent-tumor-spread/) when he was recruited together with Dr. Lyden and Dr. Y. Kang to Champalimaud Foundation (CF) (http://www.jn.pt/sociedade/interior/beleza-lanca-programa-de-investigacao-do-cancro-1227085.html). For some reason, Dr. Kalluri, later on, left CF/Harvard and went to Coimbra/MD Anderson cancer center
LikeLike
Hi Leonid
For more of the shenanigans at MD Anderson read the blog of Dr. Len Zwelling.
It’s easy to find via google.
Cheers oliver
LikeLike
Yes there is an interesting post in Dr Zwelling’s blog: the question is you why you are not anymore the right person in the right place?
LikeLike
Thanks Oliver, he even refers to my website! http://lenzwelling.blogspot.de/2017/04/tactic-or-strategy-missiles-into-syria.html
LikeLike
that’s an old version of the site.
Here is the current version of the website:
http://lenzwelling.com/
Cheers oliver
LikeLike
Also, being Dr. DePinho a luso-descendent, are very curious the connections of Dr. Kalluri not only with MD Anderson but also with research institutions in Portugal: first, with Champalimaud Foundation (ttps://indigoprojects.eu/stakeholder/5) then with University of Coimbra (http://www.cnbc.pt/research/department_group_show.asp?iddep=1222&idgrp=1253), besides with Sonia Melo and IPATIMUP
LikeLike
Another update:
Nonetheless the problematic and polemic GPC1 paper, Dr Kalluri published this month a new exosome and pancreatic cancer paper again in Nature and again this last paper already is being commented in pubpeer with similar issues to the Ines found in the GPC1 papers
LikeLike
This makes call back and remember Dr Randy Schekman, Nobel prize for medicine, about luxury journals that distort science in 2013
LikeLike
How can Nature continue to publish impossible research data from the Kalluri group?
I did never get any explanation for the immunoelectronmicroscopy results for GPC1 presented in their Nature article from 2015 (Melo SA et al.) regarding the heavy staining in the middle of the exosomes, without any staining at the membrane (where you should expect to locate it).
https://pubpeer.com/publications/70714D8ACB8F13164A2752B4335F38
In their new exosome paper published in Nature (Kamerkar S et al., 2017) they again present immunoEM data showing CD9 inside the exosome, again no staining along the membrane.
https://pubpeer.com/publications/E70D3000307B950841F82BE9CA5326
This is impossible if the staining supposed to be specific, and the shape of the vesicle also makes me think of an artifact. I wonder who where the reviewers that approved this manuscript?
LikeLike
Pingback: Yoshinori Watanabe data manipulations: much worse than officially presented – For Better Science
Pingback: Zombie scientist Sonia Melo awarded by AstraZeneca – For Better Science
Pingback: Flawed cytometry of Rector Giorgio Zauli – For Better Science
Pingback: Preprinters of the World Unite – For Better Science
Pingback: How Andrea Cerutti molested and defiled Journal of Immunology – For Better Science
Pingback: The Redemption of Arturo Casadevall – For Better Science
Pingback: Corrupción en la investigación científica, un problema estructural – Información Alternativa
Pingback: David Sabatini TORmented by steaming turds – For Better Science
Pingback: Naughty dentists, or how UCLA hunts a whistleblower – For Better Science
Pingback: Mr ACE2 Josef Penninger, Greatest Scientist of Our Time – For Better Science
Pingback: Bad Choices in Dresden – For Better Science
Pingback: Croce begat Calin, and Calin begat Girnita… – For Better Science
Pingback: Bad Choices in Dresden III – For Better Science