The Portuguese cancer researcher Sonia Melo has now achieved the status of a zombie scientist. After an internal investigation which records are kept secret, she was cleared of all suspicions of scientific misconduct and re-installed as group leader at the Instituto de Investigação e Inovação em Saúde (I3S) in Porto (see my report here). This despite an impressive PubPeer record of data integrity concerns, and despite the fact that the European research society EMBO revoked Melo’s Installation Grant funding after having determined problems with her publications. EMBO nevertheless stick to their decision, but Melo’s Portuguese funders like Fundação para a Ciência e a Tecnologia (FCT) apparently see absolutely no need to reconsider their support, certainly not after the I3S whitewashing. Melo previously had to retract a paper (Melo et al, Nature Genetics, 2009) for data manipulations, her other works were however found not problematic by the I3S commission. In two papers in Cancer Cell (Melo et al 2010 and Melo et al 2014), the alleged duplications were apparently proven not to be duplications. As I learned, this was probably because while the top part of the gel images indeed did look suspiciously similar, the lower parts were clearly different. A possibility of digital image splicing was not considered, as it seems. In any case, even if the top bands are indeed the same, it doesn’t really matter. Cell editorial offices made on several occasions perfectly clear that data integrity is not one of their top concerns.
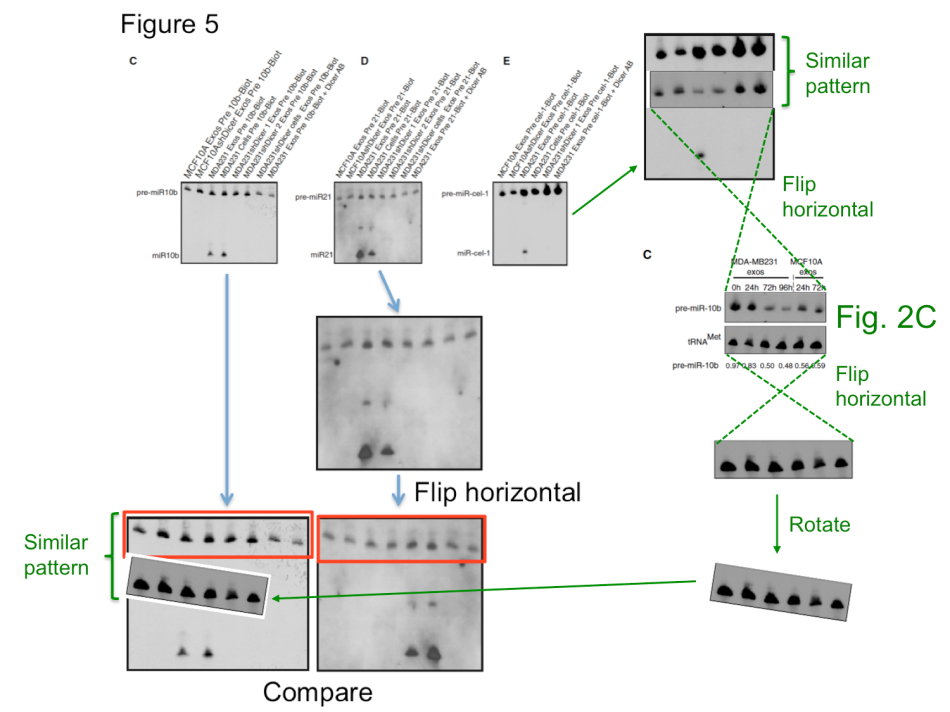
Two corrections were recommended by I3S experts, for Melo et al, PNAS 2011 and Melo et al, Nature 2015. The problem with the latter paper is however: it seems to be utterly irreproducible. This discovery of Glypican-1 protein as a specific and unique marker of early stages of pancreatic cancer to be detected in the patients’ blood was made in the lab of Melo’s former boss Raghu Kalluri at MD Anderson in Texas. It brought hope to many cancer patients, but also to biotech investors. Kalluri’s company Codiak Biosciences raised at least $80 Million in venture capital to market his and Melo’s patented discovery, with direct involvement of MD Anderson. It is reasonable to assume that the investors might have gotten second thoughts should Melo or even Kalluri be found guilty of misconduct. They might even be tempted to ask for their millions back. In this regard, one kind of understands why MD Anderson never bothered to investigate the data integrity concerns in Kalluri’s publications with Melo and also without her. It might even explain why the I3S investigation found something that different from that by EMBO, while analysing exactly the same papers by Melo.
Below Ana Pedro, a former peer of Melo, offers her post-publication peer review of that Nature 2015 publication. She shows why it is unlikely for Glypican-1 to be specifically detectable on the cancer cell exosomes (cell-membrane-derived vesicles which our cells shed into their surroundings and into blood). Pedro analysis is also supported by the fact that Melo et al discovery is primarily focused on the use of excessively high concentrations of a certain polyclonal Glypican-1 antibody which sale has been discontinued since. There is no published evidence that the central claims of the Nature 2015 paper were ever reproduced using any other Glypican-1 antibody either.
An anonymous commenter calling himself “Frank” (who soon turned out to be a senior researcher at I3S associated institution Ipatimup, where Melo is also employed) took as stand defending the investigation and Melo’s research integrity in the comment section on my site. He eventually abandoned it as his forum and apparently moved to join the mudslinging affray at PubPeer, assisted by one or more other anonymous defenders of Melo’s research. As evidence for the reproducibility of Melo’s Nature paper, “Frank” kept pointing to this publication from the nanotechnology engineering lab of Thomas Thundat at the University of Alberta, Canada: Etayash et al, Nanoscale, 2016. Another reader of my site countered:
“Regarding the publication in Nanoscale: There are no results showing specificity for the anti-GPC1 antibody they are using. Without any documentation showing antibody specificity in their cell model, the data is in my eyes worthless”.
In fact, even Thundat himself was not really defending the specificity of his assay. In his emails to me he spoke of another paper “from a different group” he saw, “about real patient samples” (which is not published yet) as well as “some very exciting results with mouse” which he was recently shown by his visitors from US, which he then admitted were “preliminary”.
To my question about the discontinued Glypican-1 antibody by ThermoFisher Thundat declared:
“The people whom I am going to collaborate with are going to send us the antibodies/receptors they developed. Our focus is developing readout technology for high sensitivity, real-time multiplexed detection. Specificity is going to come from detecting multiple biomarkers simultaneously. From experiments we have to determine the best combination of biomarkers”.
With this we probably can dismiss Thundat’s publication as proof of reproducibility of Melo et al seminal work in Nature.
What else? The Nature 2015 paper describes the comparative analysis of cancer serum samples versus those from healthy donors. The experimental methods description however does not make it entirely clear if both cancer and healthy samples were processed equally. We do learn that the cancer samples originated from the University of Heidelberg and the University Hospital of Dresden in Germany, while the paper’s last author Raghu Kalluri suggested in an prepared statement elsewhere those blood sera were up to 30 years old. I attempted to contact his and Melo’s responsible co-authors from the Dresden University, namely Juergen Weitz, Nuh Rahbari, Christoph Reissfelder, and Christian Pilarsky, with simple questions about the age, origins, preparation methodology and storage conditions of the cancer and healthy control samples. All rather reasonable questions, which should have been already addressed in their published Materials and Methods section. Especially since blood does tend to deteriorate rather quickly, which certainly calls for tightly controlled experimental conditions when comparing different sample groups. I received no reply from any of these four German clinicians. Even my reminder email was met with thundering silence. Shall we therefore assume that the healthy controls were indeed relatively fresh and the cancer samples were indeed up to 30 years old? At which temperature were they stored during that time? Were they actually any good for such a comparative exosome analysis? Does it matter to the authors that the Glypican-1 test they are now commercially developing might randomly assign false-positive pancreatic cancer diagnoses to perfectly healthy patients?
Our traditional academic culture teaches that a published paper must never be questioned, because it already was peer reviewed. Especially a paper published in the apical journal Nature, where the editorial scrutiny and peer review is supposed to be the most rigorous. As my former boss like to say to his critics: once you also published in Nature, then you can come questioning my paper there. Indeed, when doubts about published elite research arise anyway, the etiquette dictates that only elite researchers who themselves regularly publish on the same journal impact factor level are entitled to express a cautious opinion. Even then, their letters to editors often land in the trash bin.
However, to tell it back to our science elite with a Nobel-Prized quote: “the times they are a changin’”. Here is a post-publication peer review which is based on evidence, not eminence.
Is GPC1 from serum exosomes a marker to diagnose pancreatic cancer?
Ana Pedro, MPharm, PhD
United Kingdom
anacadavezpedroAThotmailDOTcom
Extracellular vesicles (EVs) are nanometer-sized membranous vesicles which are involved in cell-to-cell communication. EVs contain several types of functional molecules, such as proteins, mRNAs, and microRNAs (miRNAs). Increasing evidence suggests a key role for EV-mediated intercellular communication in a variety of cellular processes involved in tumor development and progression, including immune suppression, angiogenesis, and metastasis. Therefore, EVs are emerging as potential therapeutic targets in cancer therapy.
The publication by Melo et al., Nature 2015, claimed that presence of glypican 1 (GPC1) in serum exosomes unequivocally identifies patients with pancreatic ductal adenocarcinoma (PDAC) cancer. As PDAC has an extremely poor prognosis and currently no robust biomarkers for the early stages of the disease are available, the study by Melo et al. carried great promise. However, PubPeer commenters have raised serious concerns about the specificity of the polyclonal anti-GPC1 (ThermoFischer, catalogue PA5-28055) used in this publication. These concerns were further fuelled by the discontinuation of this antibody preparation, since the validity of the presented results depends entirely on the fidelity of those antibodies.
Considering the clinical impact of a valid PDAC marker and the grave concerns raised by parts of the scientific community on PubPeer, I here confront the data published by Melo et al. with GPC1 expression data available in several databases such as The Human Protein Atlas, PeptideAtlas, and Vesiclepedia, as well as in publications, and found important discrepancies.

GPC1 expression levels in healthy pancreas are only moderate.
By consulting The Human Protein Atlas, I found that in normal pancreas GPC1 transcripts are not highly expressed compared to other tissues (Fig. 1a) and, coherently GPC1 protein is not detected by immunohistochemistry (IHC) in this tissue (Fig. 1b). In agreement, mass spectrometry analyses summarized in PeptideAtlas for GPC1 presence in adult human tissues reveal only a modest amount of GPC1 in the pancreas (Fig. 2). Thus, consistently across databases, there are no indications that GPC1 expression in the normal pancreas is high.
GPC1 expression levels in PDAC pancreas are only moderately elevated.
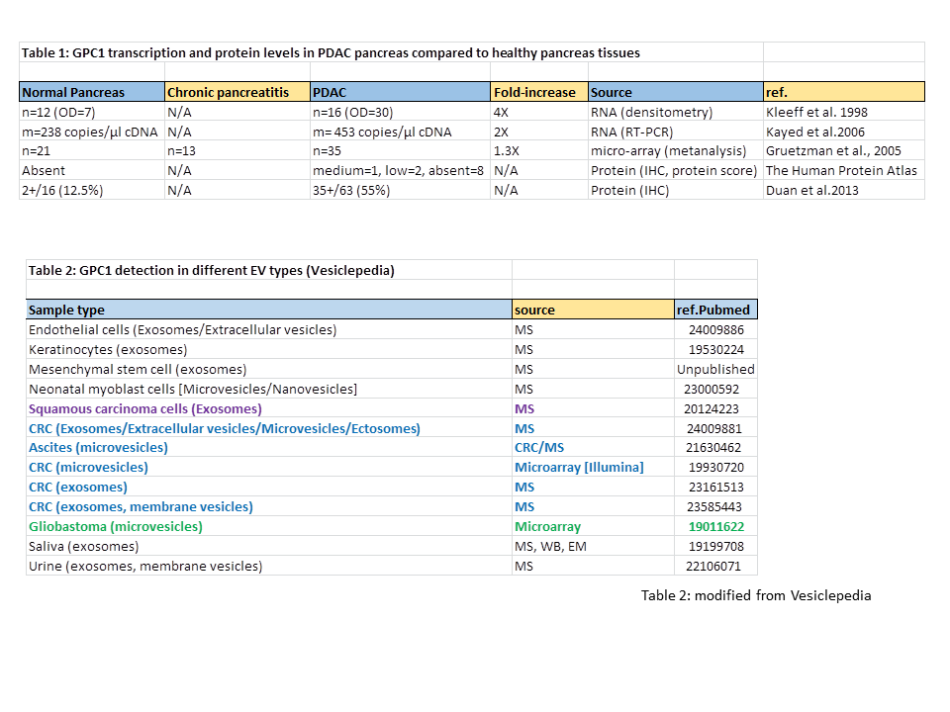
Several authors found elevated GPC1 transcription levels in PDAC pancreas compared to healthy pancreas. However, in most cases the observed increases are only few-fold (Kayed et al. 2006; Gruetzman et al., 2005; Kleeff et al., 1998), as summarized in Table 1. According to The Human Protein Atlas these increases are too modest to make pancreatic cancers sites of high GPC1 expression compared to the rest of the body, because based on IHC analysis they consider the GPC1 expression level as only “medium” in 1 of 11 samples investigated, “low” in 2 samples, and “absent” in 8 samples (quantitative scaling like for Fig. 1b). This modest upregulation is also consistent with IHC analyses by Duan et al, 2013 (Table 1) who found 2 of 16 healthy pancreases, and only 35 of 63 PDAC pancreases, to be positive for anti-GPC1 staining (Table 1).
There are no reasons to believe that GPC1 is enriched in PDAC-derived EVs.
Finally, by consulting Vesiclepedia, I found that extracellular vesicles (EV) of cancerous as well as of non-cancerous cell types may contain GPC1 (Table 2) and that the inclusion of GPC1 into exosomes is not a hallmark of a smaller degree of differentiation as, e.g., EVs of differentiated keratinocytes contain more GPC1 than EVs of undifferentiated keratinocytes (Table 1 in Chavez-Muñoz et al, 2009). The latter observation is of relevance because PDAC tumour cells are considered to be less differentiated than their healthy precursor cells. Figure 3 shows that Melo et al. could readily detect GPC1 in lysates of various cell lines by immunoblot experiments. In contrast, this analysis of the matching exosomes from those cell lines probably suffered from severe background staining (Fig. 3), arguing against relative enrichment of GPC1 in exosomes as compared to cells. There were no arguments provided to trust that the band in this immunoblot from Melo et al. (green arrow in Fig. 3) is specific and the sole indicator of GPC1 presence. However, even so, then it should be concluded that, considering the amounts present in the cell lysates, GPC1 was most efficiently incorporated in the exosomes from the non-cancerous primary human dermal fibroblasts (HDF, Fig. 3).

There are no reasons to believe that PDAC tumors release particular high numbers of EVs.
An average healthy pancreas is about 80 gram in weight, which is very small compared to the rest of the body. This weight may not be so different in the early stage of pancreatic cancer, the stage which Melo et al. suggest to be able to detect. Furthermore, PDAC tumours are poorly vascularized, and so far no evidence has been presented that these tumour cells release more EVs than other cell types. Melo et al. compared the exosomes of several non-tumour cell lines with those of pancreatic tumour cell lines, and would have noted if there were considerable differences in EV numbers. Summarized, there is no reason to believe that a sufficiently large percentage of the exosomes in sera from patients with early stages of PDAC are derived from tumour cells.
Conclusions.
Investigation of expression data in public repositories and the literature indicate that:
PDAC tumor cells:
- do not express particular high amounts of GPC1 compared to the rest of the body,
- do not selectively enrich GPC1 into their exosomes, while other cells do not selectively exclude GPC1 from their exosomes,
- do not release particular high numbers of EVs.
Consequentially the detection of GPC1 in serum exosomes is a very unlikely diagnostic marker for pancreatic cancer. Together with the serious technical concerns regarding the Melo et al. study (discussed on PubPeer), their promising message unfortunately has important unresolved issues.
Possible conflict of interest
I am an aspiring scientist who worked on exosome analysis for several tumour models. Against my desire my career was halted, and I feel that was in part because I could not reproduce several exciting messages like those of the Melo et al. study. With the here presented analyses of databases and literature I hope to show that specific detection in serum of tumour-derived exosomes based on GPC1 presence is not within expectations. I hope this will partly redeem my work, which I did with a lot of passion.




Our focus is developing readout technology for high sensitivity, real-time multiplexed detection. Specificity is going to come from detecting multiple biomarkers simultaneously.
Just like Theranos, then.
LikeLike
I have worked on Glypicans for many years, and published a large number of papers on this subject.
I have carefully looked at the issue of the specificity of the GPC1 antibody used by Melo et al., which is not available any more. However, Thermo Fisher has still images of Western blots results with this antibody (PA5-28055) on its on line catalog. For exampl,e they show a Western blot analysis of GPC1 in MDA-MB-231 cells. They show that the band corresponding to GPC1 is about 60kDa (52 after digestion with Tunicamycin). However, in the Melo paper in Extented Data Fig. 1f the band that is shown as corresponding to GPC1 runs as a 75 kDa protein (see Supplementary Figure 1f). So the band identified by the manufacturer as corresponding to GPC1 is not the same as the band indicated in the paper. I conclude that the results are not reliable and should be retracted.
LikeLike
Western blot bands size matter. Grants size matter even more.
LikeLike
[this comment was posted from an IP address identified with Fundacao para a Ciencia e a Tecnologia, I.P., Universidade do Porto. -LS]
Dear Mr. Schneider,
If your intention is to stalk forgery and falsification, I would suggest you to avoid falsehoods in your statements. Sonia Melo is not employed by the i3S and is not a group leader at the i3S. To be sure you can simply view the list of i3S groups and group leaders at the following address:http://www.i3s.up.pt/research/research-groups
It would be fair to immediately publish a “corrigendum” indicating the right Sonia Melo employer and position.
LikeLike
Hi Pat,
I don’t quite follow… Clearly says that Sonia Melo is “Principal Researcher” at the I3S “Genetic Dynamics of Cancer Cells Lab”.
Regarding employment/funding: “Our research program is fully funded and we got a 100% success rate (3 out of 3) in the 2014 FCT Call for Project Grants. Our research laboratory is organized in three intimately related research lines centered on cancer genetics, exosomes biology and cancer microenvironment.”
sources:
http://www.i3s.up.pt/research-groups/cancer/genetic-dynamics-cancer-cells
http://www.i3s.up.pt/research-groups/cancer/genetic-dynamics-cancer-cells/person/sonia-melo
LikeLike
Exactly ! Contary to what claimed by Leonid Schneider, Sonia Melo is nor Group Leader at i3S, nor employed by i3S !!
LikeLike
I don’t believe the paper either, but I do want to point out that Table 1 has some issues. PDAC is notorious for having low tumor purity, so it’s possible that the fold-change in GPC1 mRNA is underestimated by the data. For the IHC data, is it clear that the antibodies used there were really any good either? I’ve found HPA to be quite hit-or-miss…
The problems with the paper’s changing WBs band sizes are the biggest smoking gun here, in my opinion.
LikeLike
Dear R-to-R,
Thank you for your contribution to the discussion. Although PDAC is
characterized by abundant stroma, also the mass of tumour cells is
abundant and the transcript dilution effect should at most be
several-fold. Furthermore, high GPC1 expression in the tumour cells would
probably have been picked up by at least one of the IHC studies
summarized in my article. The point of Table 1, in combination with the
other figures, is that findings by multiple studies consistently find no
evidence for particularly high GPC1 expression in PDAC tumour cells.
LikeLike
Thanks for your reply. I do want to point out though that several PDAC RNA studies have seen tumor purities consistently as low as 20-30%, meaning that the 2- or 4-fold increase in PDAC GPC1 in Table 1 might actually be anywhere from 6- to 20-fold in reality. If true, I would consider that quite significant – of course I’m not saying this is really the case, just a caveat.
As for the IHC, I’m asking if any of the antibodies are really any good. From what I’ve read, the different antibodies tend to give different and confusing band patterns, so it’s not clear that ANY of the antibodies used in HPA etc are specifically detecting GPC1. If so, then absence of evidence is not evidence of absence. But, I’m not a GPC1 expert, hence my asking.
LikeLike
Dear R-to-R
My article is only about ‘absence of evidence’ for the premise in the Melo et al. study that GPC1 expression on PDAC tumor cells is unusually high, so you and I don’t disagree. You are correct in that analysis of RNA expression at the whole tissue level may underscore effects at the cell-type level, and that IHC reports deserve some caution. However, in addition to what I already wrote:
The Human Protein Atlas used anti-GPC1 from Sigma, HPA030571, whereas Duan et al. used anti-GPC1 from Santa Cruz Biotechnology (the number was not specified). Thus two independent groups, each using different antibodies, found by IHC method that PDAC cells do not express very high amounts of GPC1.
Melo et al. found only 3-5 fold more GPC1 RNA in pancreatic cancer cell lines than in the compared non-tumor cells.
Melo et al. based their premise on their reference 18, as they explain ‘Glypican-1 (GPC1) is a membrane-anchored protein that is overexpressed in breast and pancreatic cancer17–19’ and ‘Many cancer cells overexpress GPC1, with the most abundant increases observed in pancreatic cancer cells lines and tissue17–19’. Their references 17 and 19 are about breast cancer and gliomas, and their reference 18 is the study by Kleeff et al. that is included in my Table 1. I believe that my Table 1 is an objective summary of the relevant studies.
There seem to be no arguments for believing that GPC1 expression on PDAC tumor cells is unusually high, while there are several arguments against it. Researchers who do believe that it is unusually high should provide evidence for that, as the burden of proof rests on them.
Ana
LikeLike
Hi Ana, haven’t checked this in a while, appreciate your response.
However, I still have to say that your approach to the IHC data isn’t quite right. You say that “There seem to be no arguments for believing that GPC1 expression on PDAC tumor cells is unusually high, while there are several arguments against it.” but then present as arguments against it “two independent groups, each using different antibodies, found by IHC method that PDAC cells do not express very high amounts of GPC1.”
If both of the antibodies HPA030571 and Santa Cruz are not good for detecting GPC1 by IHC, then they are as good as no evidence at all, wouldn’t matter if you have 100 of them. Similar reasoning with the RNA data.
While we both agree “they have not supported their data”, that’s a VERY difference stance than saying “there are good arguments against their data.”
LikeLike
There are several problems regarding this paper. I have discussed the EM data of GPC1 in what they claim are exosomes with an expert in EM staining at our institute. He could confirm the worries I feel regarding the unusual strong staining and unusual exosome morohology. He have never experienced so strong immuno staining and the ultra structure look odd. One of the exosomes represents a perfect circle. A hole in the grid?
LikeLike
Morty,
Can you ask your man if not a lot of gold should be outside the circles? GPC1 is at the surface, right?
I don’t get the plane of focus either.
Thanks!
LikeLike
Silver, you are right. You would expect staining along the membrane, not heavy “aggregates” in the lumen.
Remember this is thin sections <100 nm thick and the number of antigens on the surface of such a thin section is very limited.
LikeLike
Morty, Thanks! So it is a section view and not a surface view? Can’t figure out. Hey, even if surface view, this would be denser than single spike viruses!
LikeLike
As a 28 year old chemist who likely has pancreatic cancer, and a bunch of negative tests, this paper represented hope for me. Realizing that, like much of academia, it is grounded in the quest for grant money and ultimately trash is a huge blow. I was just considering going to grad school because I was inspired by this work. I feel certain now that my intelligence would be wasted there.
In a way, I am glad. Because now I know that my talents will continue to be be better spent in industry. Let the fist of God come down on the university system for its dishonest ways. As more and more students become disenfranchised with the differential between what college promises and the return on their financial and intellectual investment, surely the system will collapse under its own weight. Capitalism will not solve the problems of man. But certainly it does a better job than the eggheads in their ivory towers, bilking the taxpayers of their hard-earned money to line their offices with accolades.
LikeLike
Hi Rags-to-Riches,
A recent study seems to further support my point of views relative to
the Melo et al. paper. This is what was written on PubPeer (by Peer 18):
In Lai et al., 2017 (http://www.sciencedirect.com/science/article/pii/S0304383517301283), the authors conclude that exosomal GPC1 is not diagnostic for PDCA (quite literally “We report that exosomal GPC1 is not diagnostic for PDAC”). They say that on average, perhaps, there could be a small increase in exosomal GPC1 in plasma of PDAC patients, but the authors conclude that this observation is not significant. Indeed, only a few patients samples were investigated and some healthy individuals showed higher exosomal GPC1 than detected in some PDAC patients.
Lai et al. suggest that the differences they found in comparison with the Melo et al. study may have been due to a difference in techniques. They determined the GPC1 amounts by mass spectrometry, whereas Melo et al. relied on the sensitivity of a no longer commercially available antibody preparation. Lai et al. speculate that recognition by those antibodies may have been sensitive to qualitative differences in glycanation of GPC1 glycosaminoglygans side chains in cancer cells. Theoretically, that is a possibility, but it should be considered that:
– Size differences for GPC1 in exosomes from cancer and non-cancer cells have not been reported yet, and the Western blot in Melo et al. ED Fig. 1f does not show any differences in apparent size of GPC1 in cell lysates. Their Western blot for GPC1 in exosomes (Melo et al. ED Fig. 1g) is probably not reliable enough for making any conclusions (as discussed extensively in this thread).
– Melo et al. showed similar results for exosomes in a mouse PDAC model, which would necessitate that this putative special cancer-type GPC1 epitope is the same in human and mouse,
– Melo et al. used a quite undiluted antibody preparation which when used for labeling patient exosomes exhibited unusual abundant staining with a pattern not expected for specific labeling of a membrane protein (the electron micrographs in Melo et al. Fig. 2c).
– Quite fundamentally, no proper evidence seems to have been presented that the antibodies used by Melo et al. can specifically detect non-denatured GPC1 at membrane surfaces of cells or exosomes of human and mouse.
It is also of interest that in a second recent study, by Liang et al. 2017, the authors found another exosomal marker for pancreatic cancer, EphA2, and briefly discussed the Melo et al. study but do not mention whether they investigated GPC1 themselves (http://www.nature.com/articles/s41551-016-0021). A direct comparison of EphA2 and GPC1 would have been logical and interesting.
LikeLike
Hello again, R-to-R
Even more results are supporting my point of views relative to the GPC1 paper. Another group, curiously AGAIN from MD Anderson Cancer Centre, found another set of plasma markers for pancreatic cancer as it can be read here (Balasenthil et al., 2017): https://www.sciencedaily.com/releases/2017/02/170228185217.htm
In this study, multiple markers are described what increases the possibilities of correctly detecting early stage pancreatic cancer. Briefly, “deploying TFPI and an isoform of tenascin C, TNC-FN IIIC, with CA 19-9 improves performance discriminating stage I and II disease from healthy or benign disease controls”. Isn’t it curious that even colleagues reporting on the same topic don’t refer to the Melo et al., 2015, study, in spite of the reported ROC=1?
LikeLike
Pingback: Melo and Kalluri defend discredited Nature paper with preprint, where they admit data “adjustments” – For Better Science
Pingback: Zombie scientist Sonia Melo awarded by AstraZeneca – For Better Science