Another retraction hits the fallen star plant scientist Olivier Voinnet. This time it is a previously corrected paper in Science, which was pulled after new evidence of further data manipulations appeared on PubPeer some months after its 3rd correction.
This makes it Voinnet’s 8th retraction, but the loss of the Dunoyer et al 2010 paper is more than just another brick falling from his great publishing record edifice. It is one of the two pillars of an entire school of thought in the field of plant pathogen defence, which we now witness crumbling into dust (see my earlier report and a brief summary below).
The Dunoyer et al 2010 paper has been corrected twice before (both are behind paywall, the one from April 15th 2011 acknowledges image reuse: “The image in Fig. 3B (center panel) was previously published as Fig. 1e in P. Dunoyer et al., Nat. Genet. 39, 848 (2007)”). The third correction from January 22nd 2016 declared a shopping list of further image irregularities, with Patrice Dunoyer taking full responsibility, just like he did on many other occasions (see my earlier report here).
Passing the buck
Dunoyer used to be Voinnet’s right-hand man at the CNRS Institut de Biologie Moléculaire des Plantes in Strasbourg, France. When Voinnet was delegated by CNRS to ETH Zürich to assume his professorship for as long as he desires, he took most of his junior researchers to the new Swiss lab. Yet the loyal and tenured Dunoyer remained behind, leading the master’s lab in his absence. When the avalanche of data manipulations discovered in around 40 Voinnet papers grew and came crashing down in early 2015, Dunoyer and Voinnet were both investigated by CNRS in a top-secret procedure; both were made to come and defend themselves in front of a committee in Strasbourg. Dunoyer’s loyalty and readiness to accept the entire blame seemed to pay off: though he was excluded from his CNRS position for 1 year, 11 months of that were suspended. Basically, Dunoyer lost just one month of salary. His boss however was suspended from CNRS funding for 2 years, after an earlier decree by the CNRS president Alain Fuchs redefined research fraud in a manner which precluded Voinnet from being found guilty of it and thus automatically facing a sack. This 2-year suspension however formally made Voinnet’s return to his permanent CNRS position in France impossible. The “cheater professor” lost his Strasbourg lab and was stranded in Zürich, for as long as the ETH was happy to host his tenure.
Curiously, the current Science retraction does not apportion any responsibilities to anyone. This is actually quite understandable, because if Dunoyer were to accept the sole blame yet again, what with his suspended sentence in place he might be forced to move on to explore alternative careers outside of academia. Same might even have happened to Voinnet, since he previously failed to disclose these manipulations to the rather benevolent ETH investigative committee.
Afraid to look
One of two external member of this committee, Witold Filipowicz, had been paid shortly before his investigation €10,000 into his private pocket to collaborate with Voinnet and Dunoyer, it was also Filipowicz who previously nominated Voinnet for the EMBO Gold Medal, which the latter then lost in the wake of the scandal. The following was what Filipowitz and his colleagues found while investigating Voinnet’s “illustrations” (as ETH chose to describe his original research figures) in regard to Dunoyer et al 2010:
“An extreme case was represented by the use of the same loading control in three different papers (Nat. Genetics 2007 39:848; Science 2010 328: 912; EMBO J 2012 31: 2553). This issue was aggravated by the existence of an electronic library of loading controls originating from different experiments. Upon inspection of lab notebooks for the data appearing in some of the questioned papers the commission confirmed that appropriate loading controls had actually been properly performed for each experiment at the time”.
The ETH investigative committee then concluded for the now retracted paper:
“Given that primary data exist and experiments were correctly conducted and documented, the commission does not propose retraction of the trio of papers with shared loading controls (EMBO J 2012 31:2553; Science 2010 328: 912; Nat. Genetics 2007 39: 84). Corrected figures should be submitted to the journals”.
Obviously there was much, much more. Not “just” a “library” collection of generic loading controls, used to make gels appear equally loaded (while these might have been just as well mis-loaded deliberately to achieve the appearance of certain levels of gene expression). It seems neither ETH nor CNRS investigators were willing to look too closely, possibly afraid of what else they might find. Good that others did it for them, free of charge, and posted their evidence on PubPeer for all to see.
Everyone’s best interests
Another Voinnet publication which Science previously corrected in January 2016, against explicit retraction recommendation of the ETH investigative committee, remains untouched, at least for now. The paper Deleris et al, 2006 was found to contain some very heavy and outrageous manipulations, while a reader of my site provided his own detailed analysis which revealed this paper’s methodological weaknesses even outside of manipulations (see his image analysis and summary report). The journal however remained unimpressed by any of this, the former Science Editor-in-Chief Marcia McNutt chose to defend the correction with this explanation on Twitter:
The young scientist in need of protection is apparently Angelique Deleris, since 2011 research group leader at Institut de Biologie de l’École Normale Supérieure in Paris and supported by Human Frontiers Science Program funding. To assume she knew nothing of Voinnet’s manipulations of her own research data is naïve at best, as everyone who ever published a first-author paper will be able to confirm.
The thing with the Dunoyer et al 2010 retraction
The paper was published back-to-back together with the work from the lab of Voinnet’s famous mentor and PhD advisor, Sir David Baulcombe. Both Dunoyer et al 2010 and Molnar et al., 2010 manifested the role of the short double-stranded RNA nucleotides, specifically siRNA, in the plant pathogen defence. There, locally-generated siRNA molecules were allegedly proven to be acting as a fast-spreading mobile signal, warning the entire plant of the virus intrusion (for details, read my earlier investigation). Scientists had been trying to decipher the molecular identity of this elusive mobile signal for years, until Voinnet and Baulcombe have finally solved the case. Their discovery became a scientific fact, ready to go into text books.
Was the evidence ever reproduced in other labs? My earlier attempts to interview scientists in the field (some of whom Baulcombe himself even once named to me as support witnesses) brought forth not a single declaration of reproducibility. Some admitted they trusted the discovery based on the fact alone that the Baulcombe and Voinnet papers mutually corroborated each other. Mostly, what I received as feedback was graveyard silence. Occasional critics however clearly stated that Voinnet’s discovery was not reproducible and siRNA was not at all the mobile signal. Most of these peers however did not dare to be quoted even anonymously, but Dacheng Liang, followed by his former advisor Rosemary White, were not scared. They both spoke out extensively in my earlier article.
Voinnet based his claims of siRNA being the mobile signal of plant antiviral defence also on his earlier papers, mostly co- authored by Dunoyer, which does not really make them any trustworthy. These also contain image manipulations (like Dunoyer et al, Nature Genetics 2007), one was even retracted for this reason (Dunoyer et al, EMBO J, 2010). Further support is supposed to come from the undead zombie paper Deleris et al 2006, which probably even those most sympathetic Science editors do not really trust anymore. Now, with one of the two back-to-back Science-postulated pillars from 2010 collapsing, the school of siRNA as mobile signal is being solely carried by Baulcombe’s lonely paper, without anyone in sight willing to step forward and confirm its reproducibility. Baulcombe himself did not reply to my inquiry, but one should not forget that he for years failed to notice all the data manipulations happening under his nose, and thus had to retract two of his own papers (and to correct three more) featuring his own former star-PhD student. Even Voinnet’s PhD thesis made under Baulcombe’s supervision contains manipulated figures galore, but the title-giving University of East Anglia did not seem to mind much.
Critics scared
So, this main discovery of Voinnet (and possibly that of Baulcombe as well) goes into the bin. Another one of Voinnet’s big claims in Science, that of siRNA mediated antiviral defence in mammals (Maillard et al 2013), does not seem to have sprouted much follow-up research or even evidence of reproducibility, for that matter. In fact, it had already been harshly critiqued before (tenOever, Cell Reports 2014), which earned that particular critic a public accusation from the ETH commission report of having masterminded an insidious PubPeer attack on Voinnet:
“Has there been an organized effort to discredit OV and inflate accusations against
him? OV stated that a large number of emails targeting the RNA and plant biology
community (in USA, France, and Switzerland) was sent out in late 2014/early 2015.
Indeed, two members of the commission were themselves recipients of the
anonymous alert email sent on 20.12.2014 by xxxxxxxxxxxxxx. Additionally, OV
mentioned that “although he has no proof of it, he has the strong suspicion that a
colleague with whom he had a scientific conflict at Cell Reports (9: 795 and 9: 798)
might have helped orchestrate the sudden dissection of all his scientific production”.
PLOS editorial gatekeeping
I once asked during a Twitter debate David Baltrus, plant scientist at University of Arizona, to name at least one paper from Voinnet’s own lab which in his view was still scientifically reliable. This was what Baltrus eventually replied:
Well. There was that recent preprint by none other than the original Voinnet whistle-blower Vicki Vance, plant scientist nearing retirement at University of South Carolina. She exposed Voinnet’s earlier data manipulations in public, which initially prompted heavy protest and anger from the indignant plant science community, directed against… no, not Voinnet’s manipulations. Against her whistle-blowing, which soon nevertheless caused the very first Voinnet retraction (which was accidentally again a Dunoyer et al: Plant Cell, 2004).
The aforementioned preprint from Vance lab disproved Voinnet’s discovery of the molecular mechanism of a certain viral suppressor, the corresponding peer reviewed paper was recently published in the journal Plant Physiology (Mlotshwa et al, 2016). Strange thing is: the journal PLOS Pathogens, where the criticised Voinnet’s publication Jay et al 2011 was first published and then corrected for data manipulations, was surprisingly less interested in Vance lab’s attempt to reproduce it. The submission was rejected by the PLOS Pathogens editors without even being sent out for peer review. Gatekeeping editorial function working at its best, one must admit.

A recurrent nightmare
Large swathes of the plant research community as well as journals want to finally make the Voinnet scandal a thing of the past, as a kind of an almost forgotten bad dream. The journal Cell explicitly refused to do anything about Voinnet’s suspected manipulations in his 3 papers in Cell and Molecular Cell, despite or actually exactly because of the original data being unavailable. The European Research Council ERC keeps pumping heavy funding money into Voinnet’s quasi-defunct ETH lab. Yet despite everything, the Voinnet scandal keeps popping up again and again. Like with today’s annihilation of a seminal paper in Science. This is the retraction noteretraction note published today, October 14th 2016, which I accompany with an updated list of Voinnet’s corrections and retractions:
“ON 22 JANUARY 2016, Science published an Erratum (1) to the Report “Small RNA duplexes function as mobile silencing signals between plant cells” by P. Dunoyer et al. (2), one of three papers coauthored by Dr. O. Voinnet for which Science issued Errata. However, Science has been recently informed by Dr. Voinnet that the Erratum to Dunoyer et al. does not address all the figure irregularities in the paper and that these irregularities are, in fact, extensive and inappropriate image duplications and manipulations that cannot be considered the result of mistakes.
Coauthors G. Schott, C. Himber, A. Takeda, J. C. Carrington, and O. Voinnet have supplied the following details of the manipulated figures:
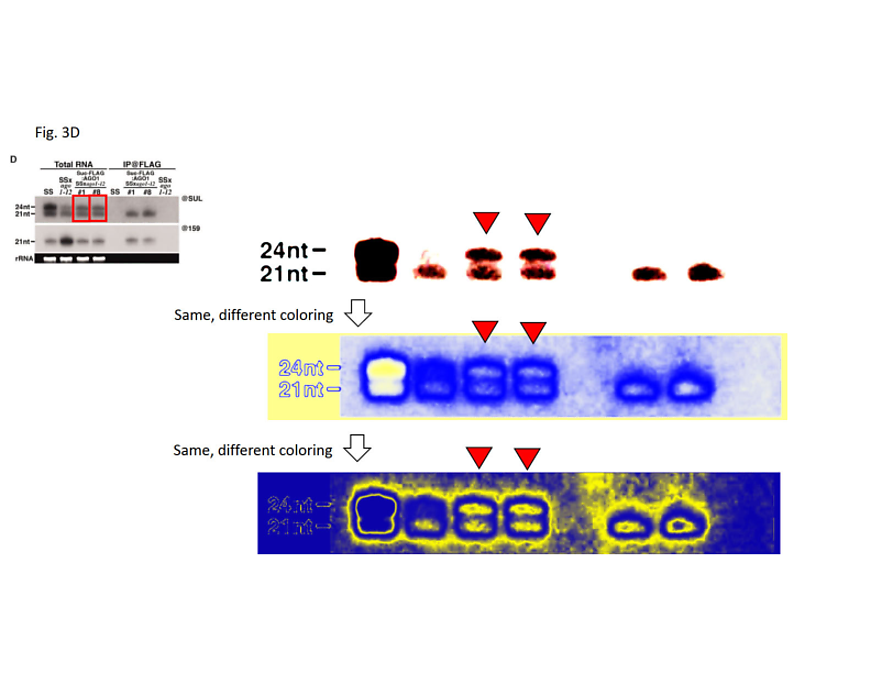
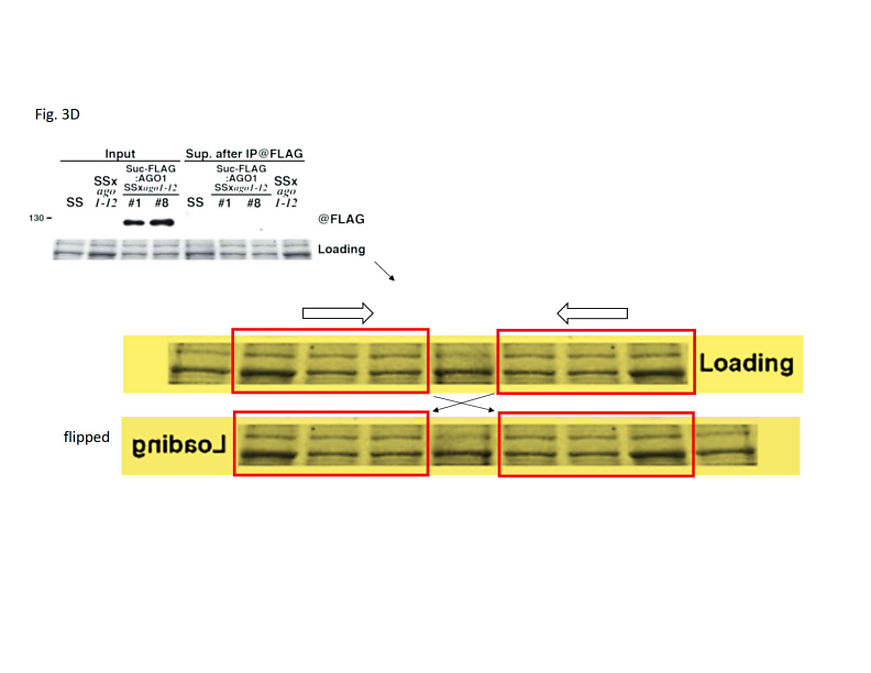
Figure 1B: The rRNA loading control was partially duplicated from an unrelated figure of a 2007 publication (3).
Figure 1D: The rRNA loading control was partially duplicated from another figure of the same 2007 study, flipped vertically and resized.
Figure 2B: The rRNA loading control was duplicated from another figure of the 2007 publication.
Figure 3D: In the bottom panel, the protein loading control has been created by inversion-duplication of an image of unknown origin. In the upper panel, the rRNA loading control was duplicated from another figure of the 2007 publication and inserted into a larger, artificial black frame to match the figure’s layout.
Figure S2C: The rRNA loading control was duplicated from another figure of the 2007 publication and inserted into a larger, artificial black frame to match the figure’s layout.
Figure S2D: The RNA loading control was duplicated from another figure of the 2007 publication and inserted into a larger, artificial black frame to match the figure’s layout. The first rRNA band on the left was bent by illegitimate image editing to match the aberrant migration pattern of the SS RNA displayed above.
Figure S3C: The rRNA loading control was duplicated from another figure of the 2007 publication and inserted into a larger, artificial black frame to match the figure’s layout.
All of the authors have agreed the paper should be retracted, except Dr. Dunoyer who did not respond to our communications. Thus, Science is retracting this paper.
Jeremy M. Berg, Editor-in-Chief
REFERENCES: 1. Science 351, aae0387 (2016); 2. P. Dunoyer, G. Schott, C. Himber, D. Meyer, A. Takeda, J. C.Carrington, O. Voinnet, Science 328, 912 (2010); 3.P. Dunoyer, C. Himber, V. Ruiz-Ferrer, A. Alioua, O. Voinnet, Nat.Genet. 39, 848 (2007)“.
RETRACTIONS:
- Brigneti G.et al.,EMBO J (1998)
- Voinnet, O.et al., Plant J. 33: 949-56 (2003)
- Dunoyer P.et al., Plant Cell 16: 1235-50 (2004)
- Moissiard G. and Voinnet O., Nat.l Acad. Sci.USA 103 (51): 19593 (2006)
- Dunoyer P.et al., EMBO J (2010)
- Dunoyer P. et al., Science 328 (5980), 912 (2010)
- Ciaudo C.et al., PLoS Genetics 9(11): e1003791 (2013)
- Sansregret R.et al., PLoS Pathogens 1(9): e1005207 (2013)
CORRECTIONS:
- Ruiz MT et al, Plant Cell10: 937–946 (1998)
- Voinnet O, et al., Nat.l Acad. Sci.USA 96:14147–14152; (1999)
- Hamilton A.et al., EMBO J (2002)
- Dunoyer P.et al., Plant J. 29: 555-67 (2002)
- Parizotto E.A.et al., Genes & Development 18: 2237-42 (2004) 2nd Correction
- Dunoyer P.et al., Nature Genetics 38: 258-63 (2006)
- Navarro L. et al., Science 312(5772): 436-439 (2006)
- Deleris A. et al, Science 313(5783): 68-71 (2006)
- Moissiard et al, RNA 13: 1268–1278 (2007)
- Haas G. et al, EMBO J 27, 2102-2112 (2008)
- Ciaudo C.et al., PLoS Genetics 5(8): e1000620 (2009)
- Bennasser Y et al., Nat. Struct. Mol. Biol. 18, 323–327 (2011)
- Jay F, et al. (2011) PLoS Pathog 7(5): e1002035
- Brodersen P.et al., Proc. Nat.l Acad. Sci. USA 109: 1778-83 (2012)
- Gibbings D. et al, Nature Cell Biology 14, 1314–1321 (2012)
- Schott G. et al, EMBO J 31, 2553-2565 (2012)
- Boccara M.,et al., PLoS Pathogens 10(1): e1003883 (2014)

Great journalism and great that the truth sometimes wins in science. Unfortunately there are many many similar cases and I hope you will continue to shed light on the dark side of science.
I never stop to be amazed by the research institutions that look another way when clear evidence of research misconduct is found. Why is not the science and discovering the truth more important than prestige and fame?
LikeLiked by 1 person
Ultimately, perhaps, the truth ALWAYS wins in science? Although it may be a tortuous process, and sometimes the untruths persist for a long time.
LikeLike
I like your optimism. Fingers crossed you are right!
LikeLike
The fragility of the scientific establishment in the face of simple, public scrutiny is amazing. No transparency, no criticism, no progress.
LikeLike
What most hurts is thinking about those patients who donate their samples thinking they will be well used in finding good treatments perhaps not for them but for future patients.
LikeLike
Some reactions from the plant science community (to be updated):
https://twitter.com/DanChitwood/status/786694037185200128
LikeLike
Great work Leonid.
Just one last question though. Are the “conclusions of the paper unaffected by these modifications.”?
LikeLike
Well, a retracted paper has no conclusions, it ceases to exist. But in this case, the conclusions were allegedly supported by a vast body of research. Only unfortunately, noone can name a specific paper or laboratory in this regard.
LikeLiked by 1 person
Proc Natl Acad Sci U S A. 2007 Sep 11;104(37):14741-6. Epub 2007 Sep 4.
Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis.
LikeLike
You seem unmoved by the number of plant scientists asserting that there is plenty of other evidence for mobile small RNAs. So, I have taken the time to amass a number of citations of work showing that small RNAs are mobile, consistent with David Baulcombe’s paper (and coincidentally, Dunoyer et al’s retracted 2010 paper).
I do not condone data manipulation or fabrication in any way, but I cannot let a headline saying “entire school of thought crashing down” stand without rebuttal.
Citations:
Martínez, G., Panda, K., Köhler, C. & Slotkin, R. K. Nature Plants 2, 16030 (2016)
Felippes, F. F. D., Ott, F. & Weigel, D. Nucleic Acids Res. 39, 2880–2889 (2011).
Carlsbecker, A. et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010
Lewsey, M. G. et al. Mobile small RNAs regulate genome-wide DNA methylation. Proc. Natl Acad. Sci. USA 113, 801–810 (2016)
Melnyk, C. W., Molnar, A. & Baulcombe, D. C. Intercellular and systemic movement of RNA silencing signals. EMBO J. 30, 3553–3563 (2011)
Calarco, J. P. et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151, 194–205 (2012)
Weiberg, A. et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013).
Miyashima, S., Koi, S., Hashimoto, T. & Nakajima, K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313 (2011)
Pant, B. D., Buhtz, A., Kehr, J. & Scheible, W.-R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738 (2008)
I hope that in the spirit of journalistic integrity you will consider correcting the assertion that this retraction has undermined a field and that the key findings are not replicable
LikeLike
Hi Ben,
as already debated over Twitter, the “school of thought” which Voinnet (and as I was told, also Baulcombe) postulated, was about siRNA being the elusive mobile signal in the long-range anti-viral gene silencing (there was some debate about the species, whether the 21nt or 24nt siRNA). This take-home message was quite clear back in 2010, greatly celebrated in several reviews, but now that the paper was retracted the research community is apparently not able to publicly explain or even agree what the paper was about (see tweets by Detlef Weigel, for example).
Of the papers you quote, some come from the Baulcombe lab, which is self-reproducibility, thus hardly the same.
I invited @JoeEcker to comment, no reply yet. So he joins the list of those scientists who have allegedly reproduced Voinnet & Baulcombe discovery but prefer not to talk about it in public.
Julia Kehr on the other hand is already quoted in my earlier report.
LikeLike
Hi Leonid
I will ask again: How long will he get to keep that Ph.D., now?
Surely, East Anglia has to budge now!
Cheers, Oliver
LikeLiked by 1 person
Hi Oliver, University of East Anglia denied my FOIA trice, so I guess this makes Voinnet for them a kind of Jesus or something. I do know for sure they were investigating, probably they cleverly stopped before finding something which would’ve forced them to act. Amen to that.
LikeLike
East Anglia also denied FOIA so consistently that it required Climategate to get the emails.
LikeLike
Copy/pasted from http://www.academie-sciences.fr/fr/Communiques-de-presse/communique-de-l-academie-des-sciences-concernant-olivier-voinnet.html :
“12 Oct 2016 – Communiqué de l’Académie des sciences concernant Olivier Voinnet
Une commission mixte CNRS-ETH Zürich examine de manière approfondie les pratiques et les démarches scientifiques d’Olivier Voinnet et de son équipe. Olivier Voinnet a été élu à l’Académie des sciences en novembre 2014 avant la mise en cause de plusieurs de ses publications. À ce jour, il n’a pas été reçu sous la Coupole, acte solennel d’intronisation de tous les membres de l’Académie des sciences. Dès que les conclusions de la commission mixte CNRS-ETH seront connues, notre Académie prendra alors les décisions nécessaires. Pendant cette période d’attente, notre Compagnie s’abstiendra de tout commentaire.”
LikeLike
Hi Leonid,
where does the evidence that “One of two external member of this committee, Witold Filipowicz, had been paid shortly before his investigation €10,000 into his private pocket to collaborate with Voinnet and Dunoyer, (…)” come from?
LikeLike
Hi Marie,
this I reported in my earlier article:
“Update 8.04.2016: The supposedly impartial Voinnet investigator Filipowicz was also a recipient of the 2014 Chaire Gutenberg at Voinnet’s own IBMP as well as neighbouring IBMC in Strassbourg. The Gutenberg Chair is financed by the Alsace Region and the Urban Community of Strasbourg with € 60,000 of which € 10,000 went to Filipowicz personally as ‘Gutenberg Prize’ and € 50,000 were awarded to his host, the teams of LabEx NetRNA of IBMP and IBMC. Coincidently, one of LabEx NetRNA teams is still headed by Voinnet’s IBMP lab keeper and key partner in data manipulation, Patrice Dunoyer”.
ETH promised to comment to me on this, but they never did.
LikeLike
The important question is, did Olivier Voinnet or Patrice Dunoyer have any say in whom the Gutenberg Prize goes to? I agree that the relationship is not completely independent, and definitely should be commented on. But it does not make Witold Filipowicz a bribe taker, as it came out in this article.
LikeLike
Marie, nobody ever insinuated a bribe here. The issue is a major conflict of interest which should have made Filipowitz unsuitable for this investigative commission. Also, prior to this, in 2008, Filipowitz had been evaluating Voinnet’s scientific performance in Strasbourg as as review board member of the Institut de Biologie Moléculaire des Plantes (IBMP). The 2014 Gutenberg award to Filipowitz could only be given thanks to IBMP, where Voinnet used to be one of its deciding leaders.
LikeLike
Thank you Leonid for clarification. I am curious about ETH’s official comment to this. Keep us posted.
LikeLike
And we still wonder why research, as an institution, is in such a profound crisis!!! Does anyone have a non-science job for me??? I will do anything that is not soooo susceptible to fraud!!!!!
LikeLike
Because of elitism, prestige and other classification garbage in science, science is becoming a ruthless jungle.
I think that a minute scrutiny of many other papers published in “so-called top” journals will reveal more striking surprises from other so-called “high-profile authors”!
Those arrogant authors who think that by publishing papers in journals C, N or S will become the scientist of all the time.
LikeLiked by 1 person
Of course what we are seeing is for sure just the tip of the iceberg…
LikeLike
Pingback: The Voinnet investigator and the tricky issue of conflict of interests – For Better Science
Pingback: The travelling circus of research integrity in Strasbourg – For Better Science
Pingback: Susana Rivas: a new research integrity scandal in French plant sciences – For Better Science
Pingback: COPE, the publishers’ Trojan horse, calls to abolish retractions – For Better Science
Pingback: Voinnet’s sidekick Dunoyer welcomed at Nature Plants, despite retractions and admitted misconduct – For Better Science
Pingback: CNRS hits back at the stream of misconduct evidence – For Better Science
Pingback: Why Nature Genetics overlooks Voinnet cheatings – For Better Science
Pingback: Olivier Voinnet: not guilty in past, present and future – For Better Science